
Culex-associated Tombus-like virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; unclassified Tombusviridae
Average proteome isoelectric point is 8.81
Get precalculated fractions of proteins
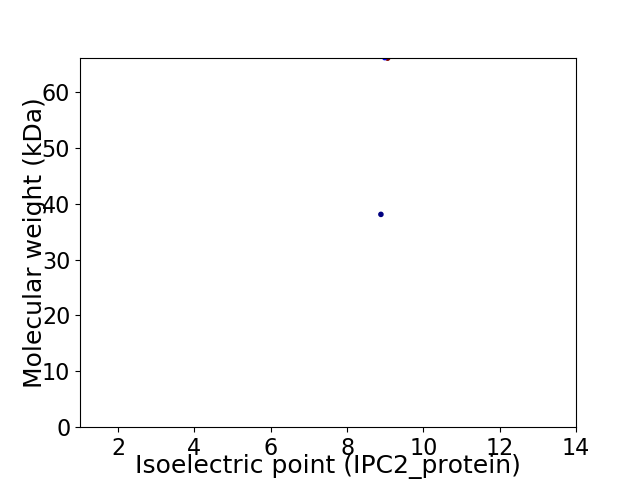
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346M220|A0A346M220_9TOMB RNA-directed RNA polymerase OS=Culex-associated Tombus-like virus OX=2304516 PE=4 SV=1
MM1 pKa = 7.54SIYY4 pKa = 10.6HH5 pKa = 6.36SMPDD9 pKa = 3.4LTRR12 pKa = 11.84PPPEE16 pKa = 5.28LGTIRR21 pKa = 11.84PIQDD25 pKa = 2.11GMMPRR30 pKa = 11.84TRR32 pKa = 11.84TQPRR36 pKa = 11.84PTGFISNLTRR46 pKa = 11.84GIQNAANRR54 pKa = 11.84AYY56 pKa = 10.69DD57 pKa = 3.38WMIPPEE63 pKa = 4.26LTDD66 pKa = 3.44AEE68 pKa = 4.43NIEE71 pKa = 4.3RR72 pKa = 11.84ADD74 pKa = 3.64EE75 pKa = 4.43VVQAWTRR82 pKa = 11.84RR83 pKa = 11.84QEE85 pKa = 4.15TEE87 pKa = 3.31IKK89 pKa = 9.78ARR91 pKa = 11.84RR92 pKa = 11.84MDD94 pKa = 3.42IDD96 pKa = 3.54NEE98 pKa = 4.42TTRR101 pKa = 11.84RR102 pKa = 11.84TAEE105 pKa = 3.75IMAQPLEE112 pKa = 4.44EE113 pKa = 4.72GLLAYY118 pKa = 10.9LNGQTRR124 pKa = 11.84GKK126 pKa = 9.75ILNKK130 pKa = 10.38DD131 pKa = 3.53YY132 pKa = 11.44VPYY135 pKa = 10.43LVKK138 pKa = 10.87LSNAYY143 pKa = 9.38FAEE146 pKa = 4.46VGQPDD151 pKa = 4.09HH152 pKa = 6.94ILRR155 pKa = 11.84EE156 pKa = 4.07KK157 pKa = 11.02VMQDD161 pKa = 3.32VIKK164 pKa = 10.64KK165 pKa = 9.9HH166 pKa = 6.25LDD168 pKa = 3.12FRR170 pKa = 11.84LKK172 pKa = 9.77DD173 pKa = 3.71TPFLTSRR180 pKa = 11.84TDD182 pKa = 2.79IMHH185 pKa = 6.69RR186 pKa = 11.84ASYY189 pKa = 10.75INALNQGKK197 pKa = 10.33RR198 pKa = 11.84ITYY201 pKa = 6.7PFWYY205 pKa = 9.79FGRR208 pKa = 11.84PKK210 pKa = 10.55VEE212 pKa = 5.1TIQEE216 pKa = 3.81WSEE219 pKa = 4.24NEE221 pKa = 3.92SGPYY225 pKa = 9.89DD226 pKa = 3.52SPRR229 pKa = 11.84PYY231 pKa = 10.24SYY233 pKa = 11.4SRR235 pKa = 11.84IILTTMGWVAVGWTAKK251 pKa = 10.6YY252 pKa = 10.33LISRR256 pKa = 11.84LYY258 pKa = 11.34SNLTSGTTTQLAVKK272 pKa = 9.46QLPLPSGTGIFRR284 pKa = 11.84KK285 pKa = 9.42FFPSTTPQVSTNSTDD300 pKa = 3.15TSGFTAYY307 pKa = 9.49ILEE310 pKa = 4.21YY311 pKa = 11.04GPIKK315 pKa = 10.83SMLVACSRR323 pKa = 11.84PVKK326 pKa = 10.37EE327 pKa = 5.07FATKK331 pKa = 10.65LPP333 pKa = 3.86
MM1 pKa = 7.54SIYY4 pKa = 10.6HH5 pKa = 6.36SMPDD9 pKa = 3.4LTRR12 pKa = 11.84PPPEE16 pKa = 5.28LGTIRR21 pKa = 11.84PIQDD25 pKa = 2.11GMMPRR30 pKa = 11.84TRR32 pKa = 11.84TQPRR36 pKa = 11.84PTGFISNLTRR46 pKa = 11.84GIQNAANRR54 pKa = 11.84AYY56 pKa = 10.69DD57 pKa = 3.38WMIPPEE63 pKa = 4.26LTDD66 pKa = 3.44AEE68 pKa = 4.43NIEE71 pKa = 4.3RR72 pKa = 11.84ADD74 pKa = 3.64EE75 pKa = 4.43VVQAWTRR82 pKa = 11.84RR83 pKa = 11.84QEE85 pKa = 4.15TEE87 pKa = 3.31IKK89 pKa = 9.78ARR91 pKa = 11.84RR92 pKa = 11.84MDD94 pKa = 3.42IDD96 pKa = 3.54NEE98 pKa = 4.42TTRR101 pKa = 11.84RR102 pKa = 11.84TAEE105 pKa = 3.75IMAQPLEE112 pKa = 4.44EE113 pKa = 4.72GLLAYY118 pKa = 10.9LNGQTRR124 pKa = 11.84GKK126 pKa = 9.75ILNKK130 pKa = 10.38DD131 pKa = 3.53YY132 pKa = 11.44VPYY135 pKa = 10.43LVKK138 pKa = 10.87LSNAYY143 pKa = 9.38FAEE146 pKa = 4.46VGQPDD151 pKa = 4.09HH152 pKa = 6.94ILRR155 pKa = 11.84EE156 pKa = 4.07KK157 pKa = 11.02VMQDD161 pKa = 3.32VIKK164 pKa = 10.64KK165 pKa = 9.9HH166 pKa = 6.25LDD168 pKa = 3.12FRR170 pKa = 11.84LKK172 pKa = 9.77DD173 pKa = 3.71TPFLTSRR180 pKa = 11.84TDD182 pKa = 2.79IMHH185 pKa = 6.69RR186 pKa = 11.84ASYY189 pKa = 10.75INALNQGKK197 pKa = 10.33RR198 pKa = 11.84ITYY201 pKa = 6.7PFWYY205 pKa = 9.79FGRR208 pKa = 11.84PKK210 pKa = 10.55VEE212 pKa = 5.1TIQEE216 pKa = 3.81WSEE219 pKa = 4.24NEE221 pKa = 3.92SGPYY225 pKa = 9.89DD226 pKa = 3.52SPRR229 pKa = 11.84PYY231 pKa = 10.24SYY233 pKa = 11.4SRR235 pKa = 11.84IILTTMGWVAVGWTAKK251 pKa = 10.6YY252 pKa = 10.33LISRR256 pKa = 11.84LYY258 pKa = 11.34SNLTSGTTTQLAVKK272 pKa = 9.46QLPLPSGTGIFRR284 pKa = 11.84KK285 pKa = 9.42FFPSTTPQVSTNSTDD300 pKa = 3.15TSGFTAYY307 pKa = 9.49ILEE310 pKa = 4.21YY311 pKa = 11.04GPIKK315 pKa = 10.83SMLVACSRR323 pKa = 11.84PVKK326 pKa = 10.37EE327 pKa = 5.07FATKK331 pKa = 10.65LPP333 pKa = 3.86
Molecular weight: 38.1 kDa
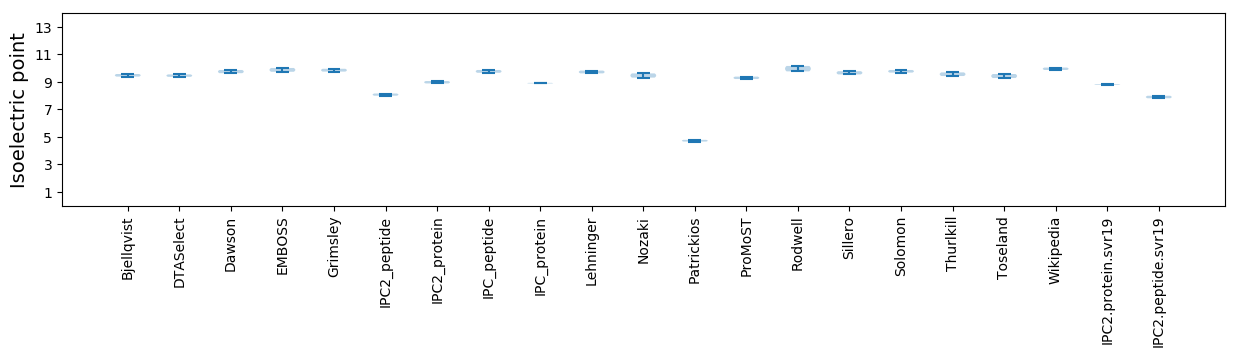
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346M220|A0A346M220_9TOMB RNA-directed RNA polymerase OS=Culex-associated Tombus-like virus OX=2304516 PE=4 SV=1
MM1 pKa = 6.49VGKK4 pKa = 10.58RR5 pKa = 11.84IGPLRR10 pKa = 11.84FTKK13 pKa = 9.92TVQLLQNYY21 pKa = 7.38PDD23 pKa = 3.52NYY25 pKa = 10.63GLGRR29 pKa = 11.84CRR31 pKa = 11.84MDD33 pKa = 3.03RR34 pKa = 11.84QIPYY38 pKa = 9.7FPSLLKK44 pKa = 10.23PDD46 pKa = 3.03EE47 pKa = 4.18WYY49 pKa = 10.41YY50 pKa = 11.42YY51 pKa = 9.19PVSSEE56 pKa = 3.73TTAAALRR63 pKa = 11.84NRR65 pKa = 11.84HH66 pKa = 5.75LPEE69 pKa = 4.42ILPEE73 pKa = 3.83YY74 pKa = 10.32DD75 pKa = 2.84AAGINKK81 pKa = 9.63LYY83 pKa = 10.57RR84 pKa = 11.84HH85 pKa = 5.41IRR87 pKa = 11.84VYY89 pKa = 10.27RR90 pKa = 11.84VHH92 pKa = 6.32PRR94 pKa = 11.84VWTHH98 pKa = 4.77QEE100 pKa = 4.14YY101 pKa = 10.13VGSMQPPSKK110 pKa = 10.26RR111 pKa = 11.84ICYY114 pKa = 9.1QAALDD119 pKa = 3.93DD120 pKa = 4.23YY121 pKa = 11.14YY122 pKa = 11.53RR123 pKa = 11.84DD124 pKa = 3.77GLISTTLQPFTKK136 pKa = 9.28IEE138 pKa = 3.81KK139 pKa = 9.3MKK141 pKa = 10.17NSKK144 pKa = 10.29YY145 pKa = 9.98KK146 pKa = 10.62APRR149 pKa = 11.84LIQARR154 pKa = 11.84KK155 pKa = 8.43PIFNIMYY162 pKa = 9.45GRR164 pKa = 11.84YY165 pKa = 9.03VKK167 pKa = 10.59ALEE170 pKa = 4.92HH171 pKa = 6.73ILFKK175 pKa = 10.27HH176 pKa = 6.17HH177 pKa = 7.13RR178 pKa = 11.84LTYY181 pKa = 10.48HH182 pKa = 6.56FGKK185 pKa = 8.9GTNEE189 pKa = 3.96NIADD193 pKa = 4.43KK194 pKa = 10.31IYY196 pKa = 10.93KK197 pKa = 10.36LSQKK201 pKa = 7.73WQWKK205 pKa = 7.85TEE207 pKa = 4.11GDD209 pKa = 3.63HH210 pKa = 6.09KK211 pKa = 10.01TFDD214 pKa = 3.08AHH216 pKa = 5.38VTVEE220 pKa = 4.11HH221 pKa = 6.75KK222 pKa = 10.86KK223 pKa = 10.7LFNKK227 pKa = 8.27FLCACFPQNVKK238 pKa = 10.13EE239 pKa = 4.2IKK241 pKa = 10.61NIGKK245 pKa = 9.77RR246 pKa = 11.84LLTAKK251 pKa = 10.13CRR253 pKa = 11.84TRR255 pKa = 11.84SGEE258 pKa = 3.7TWTVKK263 pKa = 9.07GTQQSGEE270 pKa = 4.03VDD272 pKa = 3.29TSISNTIINIAILKK286 pKa = 9.89EE287 pKa = 4.0LMHH290 pKa = 6.05QLGIKK295 pKa = 10.63GEE297 pKa = 4.17VLANGDD303 pKa = 4.11DD304 pKa = 5.18FILFTNKK311 pKa = 9.75PVDD314 pKa = 3.31IEE316 pKa = 4.11KK317 pKa = 10.82SKK319 pKa = 11.14EE320 pKa = 3.61ILKK323 pKa = 8.99TMNMEE328 pKa = 4.2TEE330 pKa = 4.11MHH332 pKa = 6.54KK333 pKa = 9.42STRR336 pKa = 11.84NIHH339 pKa = 4.95TVEE342 pKa = 4.61FCANKK347 pKa = 10.12LAFSSTGQHH356 pKa = 6.09VLFKK360 pKa = 10.62DD361 pKa = 2.93IDD363 pKa = 4.1RR364 pKa = 11.84IYY366 pKa = 11.33SKK368 pKa = 11.13FGMTNVQVDD377 pKa = 3.99YY378 pKa = 11.13YY379 pKa = 9.69RR380 pKa = 11.84TYY382 pKa = 11.57LMEE385 pKa = 4.68CAHH388 pKa = 6.62GNWKK392 pKa = 8.82MMKK395 pKa = 8.73DD396 pKa = 3.46TPIGQEE402 pKa = 3.84FKK404 pKa = 10.6RR405 pKa = 11.84IYY407 pKa = 10.65YY408 pKa = 9.33YY409 pKa = 10.15ILYY412 pKa = 10.37LEE414 pKa = 4.49EE415 pKa = 4.22QLTGQDD421 pKa = 2.93QRR423 pKa = 11.84QVQRR427 pKa = 11.84QHH429 pKa = 5.84MKK431 pKa = 10.25LQYY434 pKa = 10.39KK435 pKa = 9.69YY436 pKa = 10.9LEE438 pKa = 4.34KK439 pKa = 10.23EE440 pKa = 3.82YY441 pKa = 11.1QRR443 pKa = 11.84IIKK446 pKa = 9.11EE447 pKa = 4.03NKK449 pKa = 8.43DD450 pKa = 3.28AKK452 pKa = 10.57EE453 pKa = 4.02NTSTEE458 pKa = 4.21LTVSMMEE465 pKa = 4.7AYY467 pKa = 10.29DD468 pKa = 3.69EE469 pKa = 4.81ALDD472 pKa = 3.96LKK474 pKa = 10.83KK475 pKa = 10.26YY476 pKa = 8.1TSKK479 pKa = 11.13LINRR483 pKa = 11.84IKK485 pKa = 10.62NIYY488 pKa = 8.48TNYY491 pKa = 10.09PYY493 pKa = 9.97MGSIGLKK500 pKa = 8.17YY501 pKa = 10.11LAYY504 pKa = 10.28NSIMTISHH512 pKa = 6.35DD513 pKa = 3.64TKK515 pKa = 9.86QSEE518 pKa = 4.11RR519 pKa = 11.84HH520 pKa = 6.07KK521 pKa = 10.85YY522 pKa = 8.83TDD524 pKa = 3.24TDD526 pKa = 3.62EE527 pKa = 4.91LKK529 pKa = 10.91QSIQPLLHH537 pKa = 6.04QAFKK541 pKa = 10.52TKK543 pKa = 10.4NNRR546 pKa = 11.84HH547 pKa = 5.74IYY549 pKa = 9.83KK550 pKa = 10.29KK551 pKa = 9.84IRR553 pKa = 11.84RR554 pKa = 11.84LTGLKK559 pKa = 10.19
MM1 pKa = 6.49VGKK4 pKa = 10.58RR5 pKa = 11.84IGPLRR10 pKa = 11.84FTKK13 pKa = 9.92TVQLLQNYY21 pKa = 7.38PDD23 pKa = 3.52NYY25 pKa = 10.63GLGRR29 pKa = 11.84CRR31 pKa = 11.84MDD33 pKa = 3.03RR34 pKa = 11.84QIPYY38 pKa = 9.7FPSLLKK44 pKa = 10.23PDD46 pKa = 3.03EE47 pKa = 4.18WYY49 pKa = 10.41YY50 pKa = 11.42YY51 pKa = 9.19PVSSEE56 pKa = 3.73TTAAALRR63 pKa = 11.84NRR65 pKa = 11.84HH66 pKa = 5.75LPEE69 pKa = 4.42ILPEE73 pKa = 3.83YY74 pKa = 10.32DD75 pKa = 2.84AAGINKK81 pKa = 9.63LYY83 pKa = 10.57RR84 pKa = 11.84HH85 pKa = 5.41IRR87 pKa = 11.84VYY89 pKa = 10.27RR90 pKa = 11.84VHH92 pKa = 6.32PRR94 pKa = 11.84VWTHH98 pKa = 4.77QEE100 pKa = 4.14YY101 pKa = 10.13VGSMQPPSKK110 pKa = 10.26RR111 pKa = 11.84ICYY114 pKa = 9.1QAALDD119 pKa = 3.93DD120 pKa = 4.23YY121 pKa = 11.14YY122 pKa = 11.53RR123 pKa = 11.84DD124 pKa = 3.77GLISTTLQPFTKK136 pKa = 9.28IEE138 pKa = 3.81KK139 pKa = 9.3MKK141 pKa = 10.17NSKK144 pKa = 10.29YY145 pKa = 9.98KK146 pKa = 10.62APRR149 pKa = 11.84LIQARR154 pKa = 11.84KK155 pKa = 8.43PIFNIMYY162 pKa = 9.45GRR164 pKa = 11.84YY165 pKa = 9.03VKK167 pKa = 10.59ALEE170 pKa = 4.92HH171 pKa = 6.73ILFKK175 pKa = 10.27HH176 pKa = 6.17HH177 pKa = 7.13RR178 pKa = 11.84LTYY181 pKa = 10.48HH182 pKa = 6.56FGKK185 pKa = 8.9GTNEE189 pKa = 3.96NIADD193 pKa = 4.43KK194 pKa = 10.31IYY196 pKa = 10.93KK197 pKa = 10.36LSQKK201 pKa = 7.73WQWKK205 pKa = 7.85TEE207 pKa = 4.11GDD209 pKa = 3.63HH210 pKa = 6.09KK211 pKa = 10.01TFDD214 pKa = 3.08AHH216 pKa = 5.38VTVEE220 pKa = 4.11HH221 pKa = 6.75KK222 pKa = 10.86KK223 pKa = 10.7LFNKK227 pKa = 8.27FLCACFPQNVKK238 pKa = 10.13EE239 pKa = 4.2IKK241 pKa = 10.61NIGKK245 pKa = 9.77RR246 pKa = 11.84LLTAKK251 pKa = 10.13CRR253 pKa = 11.84TRR255 pKa = 11.84SGEE258 pKa = 3.7TWTVKK263 pKa = 9.07GTQQSGEE270 pKa = 4.03VDD272 pKa = 3.29TSISNTIINIAILKK286 pKa = 9.89EE287 pKa = 4.0LMHH290 pKa = 6.05QLGIKK295 pKa = 10.63GEE297 pKa = 4.17VLANGDD303 pKa = 4.11DD304 pKa = 5.18FILFTNKK311 pKa = 9.75PVDD314 pKa = 3.31IEE316 pKa = 4.11KK317 pKa = 10.82SKK319 pKa = 11.14EE320 pKa = 3.61ILKK323 pKa = 8.99TMNMEE328 pKa = 4.2TEE330 pKa = 4.11MHH332 pKa = 6.54KK333 pKa = 9.42STRR336 pKa = 11.84NIHH339 pKa = 4.95TVEE342 pKa = 4.61FCANKK347 pKa = 10.12LAFSSTGQHH356 pKa = 6.09VLFKK360 pKa = 10.62DD361 pKa = 2.93IDD363 pKa = 4.1RR364 pKa = 11.84IYY366 pKa = 11.33SKK368 pKa = 11.13FGMTNVQVDD377 pKa = 3.99YY378 pKa = 11.13YY379 pKa = 9.69RR380 pKa = 11.84TYY382 pKa = 11.57LMEE385 pKa = 4.68CAHH388 pKa = 6.62GNWKK392 pKa = 8.82MMKK395 pKa = 8.73DD396 pKa = 3.46TPIGQEE402 pKa = 3.84FKK404 pKa = 10.6RR405 pKa = 11.84IYY407 pKa = 10.65YY408 pKa = 9.33YY409 pKa = 10.15ILYY412 pKa = 10.37LEE414 pKa = 4.49EE415 pKa = 4.22QLTGQDD421 pKa = 2.93QRR423 pKa = 11.84QVQRR427 pKa = 11.84QHH429 pKa = 5.84MKK431 pKa = 10.25LQYY434 pKa = 10.39KK435 pKa = 9.69YY436 pKa = 10.9LEE438 pKa = 4.34KK439 pKa = 10.23EE440 pKa = 3.82YY441 pKa = 11.1QRR443 pKa = 11.84IIKK446 pKa = 9.11EE447 pKa = 4.03NKK449 pKa = 8.43DD450 pKa = 3.28AKK452 pKa = 10.57EE453 pKa = 4.02NTSTEE458 pKa = 4.21LTVSMMEE465 pKa = 4.7AYY467 pKa = 10.29DD468 pKa = 3.69EE469 pKa = 4.81ALDD472 pKa = 3.96LKK474 pKa = 10.83KK475 pKa = 10.26YY476 pKa = 8.1TSKK479 pKa = 11.13LINRR483 pKa = 11.84IKK485 pKa = 10.62NIYY488 pKa = 8.48TNYY491 pKa = 10.09PYY493 pKa = 9.97MGSIGLKK500 pKa = 8.17YY501 pKa = 10.11LAYY504 pKa = 10.28NSIMTISHH512 pKa = 6.35DD513 pKa = 3.64TKK515 pKa = 9.86QSEE518 pKa = 4.11RR519 pKa = 11.84HH520 pKa = 6.07KK521 pKa = 10.85YY522 pKa = 8.83TDD524 pKa = 3.24TDD526 pKa = 3.62EE527 pKa = 4.91LKK529 pKa = 10.91QSIQPLLHH537 pKa = 6.04QAFKK541 pKa = 10.52TKK543 pKa = 10.4NNRR546 pKa = 11.84HH547 pKa = 5.74IYY549 pKa = 9.83KK550 pKa = 10.29KK551 pKa = 9.84IRR553 pKa = 11.84RR554 pKa = 11.84LTGLKK559 pKa = 10.19
Molecular weight: 66.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
892 |
333 |
559 |
446.0 |
52.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.933 ± 0.581 | 0.897 ± 0.323 |
4.484 ± 0.011 | 5.83 ± 0.067 |
3.251 ± 0.028 | 4.933 ± 0.256 |
2.803 ± 0.868 | 7.287 ± 0.206 |
8.408 ± 1.952 | 8.184 ± 0.204 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.251 ± 0.028 | 4.596 ± 0.375 |
5.045 ± 1.497 | 4.821 ± 0.334 |
6.502 ± 0.707 | 5.157 ± 0.623 |
8.296 ± 0.874 | 3.924 ± 0.152 |
1.345 ± 0.247 | 6.054 ± 0.677 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
