
Flavobacteriaceae bacterium 144Ye
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; unclassified Flavobacteriaceae
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
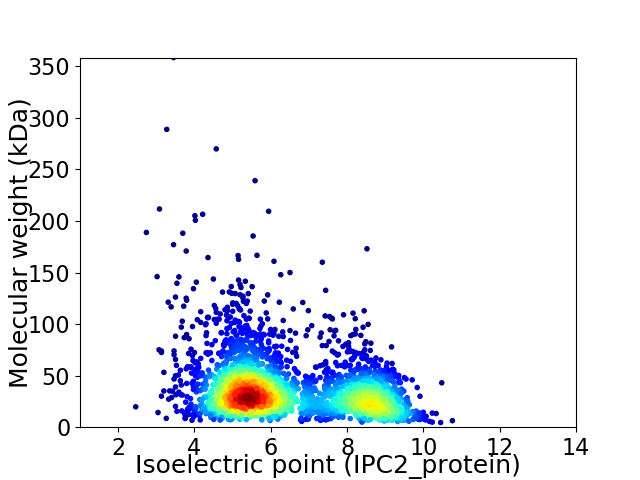
Virtual 2D-PAGE plot for 2918 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1VEB6|A0A4V1VEB6_9FLAO Uncharacterized protein OS=Flavobacteriaceae bacterium 144Ye OX=2510794 GN=EVU94_03760 PE=4 SV=1
MM1 pKa = 7.42YY2 pKa = 10.41LGQLTTYY9 pKa = 11.49NNMKK13 pKa = 10.0KK14 pKa = 10.5NIMKK18 pKa = 10.19VKK20 pKa = 9.88YY21 pKa = 8.68IWLLAVLLGFTACEE35 pKa = 4.0NDD37 pKa = 3.87DD38 pKa = 4.29DD39 pKa = 4.72SSNGGGMEE47 pKa = 4.71LPALTAGEE55 pKa = 4.48ADD57 pKa = 3.75FSNYY61 pKa = 8.93VSVGNSLTAGFSDD74 pKa = 3.31NALFIASQEE83 pKa = 3.92NSLPNILATQFAFAGGGSFTQPLMNDD109 pKa = 3.65NIGGLILGGTPVFHH123 pKa = 7.55PLTGEE128 pKa = 3.89QLFKK132 pKa = 10.77PRR134 pKa = 11.84LVTTGGAPQDD144 pKa = 4.01LEE146 pKa = 5.03DD147 pKa = 4.16VIGPVTSTTDD157 pKa = 3.58FLLNNPTGPFNNMGVPGAASFHH179 pKa = 6.72LIAPGYY185 pKa = 10.31GNIANFPAAANPYY198 pKa = 8.57FIRR201 pKa = 11.84MTGATPDD208 pKa = 3.4ASVLEE213 pKa = 4.32LAMAQSPTFFTLWAGNNDD231 pKa = 3.25VLGYY235 pKa = 8.57ATSGGDD241 pKa = 3.38GSSAITDD248 pKa = 3.23QATFDD253 pKa = 3.78FAINTLVTTLTSGGAKK269 pKa = 9.52GVMANIPDD277 pKa = 3.89VTSIPHH283 pKa = 5.86FTTVPHH289 pKa = 6.61NPLDD293 pKa = 3.8PTNEE297 pKa = 3.98DD298 pKa = 3.81FGPQIPLLNSIFGAINQIYY317 pKa = 8.97VALGQPEE324 pKa = 4.03RR325 pKa = 11.84AIVFSEE331 pKa = 4.58TEE333 pKa = 3.62ASAVVIKK340 pKa = 10.74DD341 pKa = 3.41EE342 pKa = 4.33SLDD345 pKa = 4.58DD346 pKa = 3.67ISAQITGALMASPTFPAFIAQFGLDD371 pKa = 3.62GSNPIVVQLVANLLGQTYY389 pKa = 9.56GQTRR393 pKa = 11.84QATEE397 pKa = 3.41NDD399 pKa = 3.89LFVLPSSSVIGEE411 pKa = 4.04VNMDD415 pKa = 3.51YY416 pKa = 11.44AGFLAGQGLPAEE428 pKa = 4.57LAGQFAIEE436 pKa = 5.6GITLPLEE443 pKa = 4.76DD444 pKa = 4.55KK445 pKa = 10.16WVLIPTEE452 pKa = 3.86QAEE455 pKa = 4.49IASATASFNATLEE468 pKa = 4.22TAATSAGLAFVDD480 pKa = 4.54ANSLMMQLASSGYY493 pKa = 9.35SDD495 pKa = 4.97GDD497 pKa = 3.58FTLTSDD503 pKa = 4.47LVTGGAFSLDD513 pKa = 3.36GVHH516 pKa = 5.99PTSRR520 pKa = 11.84GYY522 pKa = 11.36ALLANEE528 pKa = 4.15FMKK531 pKa = 11.1AIDD534 pKa = 3.51ATYY537 pKa = 10.49GSNFEE542 pKa = 4.27ASGNLVNIGNYY553 pKa = 5.75PTNYY557 pKa = 10.2SPTLQQ562 pKa = 3.54
MM1 pKa = 7.42YY2 pKa = 10.41LGQLTTYY9 pKa = 11.49NNMKK13 pKa = 10.0KK14 pKa = 10.5NIMKK18 pKa = 10.19VKK20 pKa = 9.88YY21 pKa = 8.68IWLLAVLLGFTACEE35 pKa = 4.0NDD37 pKa = 3.87DD38 pKa = 4.29DD39 pKa = 4.72SSNGGGMEE47 pKa = 4.71LPALTAGEE55 pKa = 4.48ADD57 pKa = 3.75FSNYY61 pKa = 8.93VSVGNSLTAGFSDD74 pKa = 3.31NALFIASQEE83 pKa = 3.92NSLPNILATQFAFAGGGSFTQPLMNDD109 pKa = 3.65NIGGLILGGTPVFHH123 pKa = 7.55PLTGEE128 pKa = 3.89QLFKK132 pKa = 10.77PRR134 pKa = 11.84LVTTGGAPQDD144 pKa = 4.01LEE146 pKa = 5.03DD147 pKa = 4.16VIGPVTSTTDD157 pKa = 3.58FLLNNPTGPFNNMGVPGAASFHH179 pKa = 6.72LIAPGYY185 pKa = 10.31GNIANFPAAANPYY198 pKa = 8.57FIRR201 pKa = 11.84MTGATPDD208 pKa = 3.4ASVLEE213 pKa = 4.32LAMAQSPTFFTLWAGNNDD231 pKa = 3.25VLGYY235 pKa = 8.57ATSGGDD241 pKa = 3.38GSSAITDD248 pKa = 3.23QATFDD253 pKa = 3.78FAINTLVTTLTSGGAKK269 pKa = 9.52GVMANIPDD277 pKa = 3.89VTSIPHH283 pKa = 5.86FTTVPHH289 pKa = 6.61NPLDD293 pKa = 3.8PTNEE297 pKa = 3.98DD298 pKa = 3.81FGPQIPLLNSIFGAINQIYY317 pKa = 8.97VALGQPEE324 pKa = 4.03RR325 pKa = 11.84AIVFSEE331 pKa = 4.58TEE333 pKa = 3.62ASAVVIKK340 pKa = 10.74DD341 pKa = 3.41EE342 pKa = 4.33SLDD345 pKa = 4.58DD346 pKa = 3.67ISAQITGALMASPTFPAFIAQFGLDD371 pKa = 3.62GSNPIVVQLVANLLGQTYY389 pKa = 9.56GQTRR393 pKa = 11.84QATEE397 pKa = 3.41NDD399 pKa = 3.89LFVLPSSSVIGEE411 pKa = 4.04VNMDD415 pKa = 3.51YY416 pKa = 11.44AGFLAGQGLPAEE428 pKa = 4.57LAGQFAIEE436 pKa = 5.6GITLPLEE443 pKa = 4.76DD444 pKa = 4.55KK445 pKa = 10.16WVLIPTEE452 pKa = 3.86QAEE455 pKa = 4.49IASATASFNATLEE468 pKa = 4.22TAATSAGLAFVDD480 pKa = 4.54ANSLMMQLASSGYY493 pKa = 9.35SDD495 pKa = 4.97GDD497 pKa = 3.58FTLTSDD503 pKa = 4.47LVTGGAFSLDD513 pKa = 3.36GVHH516 pKa = 5.99PTSRR520 pKa = 11.84GYY522 pKa = 11.36ALLANEE528 pKa = 4.15FMKK531 pKa = 11.1AIDD534 pKa = 3.51ATYY537 pKa = 10.49GSNFEE542 pKa = 4.27ASGNLVNIGNYY553 pKa = 5.75PTNYY557 pKa = 10.2SPTLQQ562 pKa = 3.54
Molecular weight: 58.71 kDa
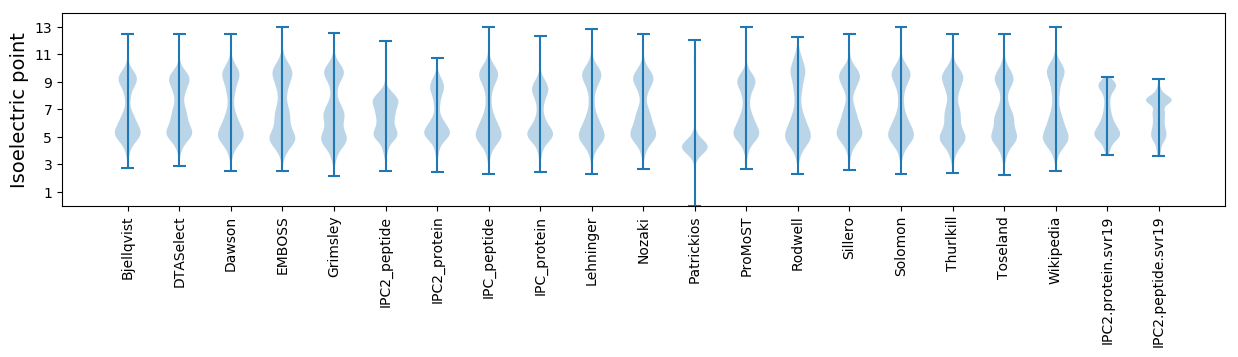
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q4B4U3|A0A4Q4B4U3_9FLAO O-succinylhomoserine sulfhydrylase OS=Flavobacteriaceae bacterium 144Ye OX=2510794 GN=metZ PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.19RR22 pKa = 11.84MATANGRR29 pKa = 11.84KK30 pKa = 8.93VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.6LSVSSEE48 pKa = 4.04SRR50 pKa = 11.84HH51 pKa = 5.63KK52 pKa = 10.69KK53 pKa = 9.55
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.19RR22 pKa = 11.84MATANGRR29 pKa = 11.84KK30 pKa = 8.93VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.6LSVSSEE48 pKa = 4.04SRR50 pKa = 11.84HH51 pKa = 5.63KK52 pKa = 10.69KK53 pKa = 9.55
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1003851 |
38 |
3435 |
344.0 |
38.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.245 ± 0.04 | 0.777 ± 0.016 |
5.825 ± 0.036 | 6.573 ± 0.042 |
5.193 ± 0.036 | 6.135 ± 0.05 |
1.786 ± 0.023 | 7.826 ± 0.043 |
7.682 ± 0.074 | 9.156 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.12 ± 0.024 | 6.496 ± 0.044 |
3.262 ± 0.026 | 3.411 ± 0.025 |
3.169 ± 0.034 | 6.601 ± 0.04 |
6.155 ± 0.064 | 6.411 ± 0.036 |
1.003 ± 0.017 | 4.176 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
