
Lytechinus variegatus variable sea urchin associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
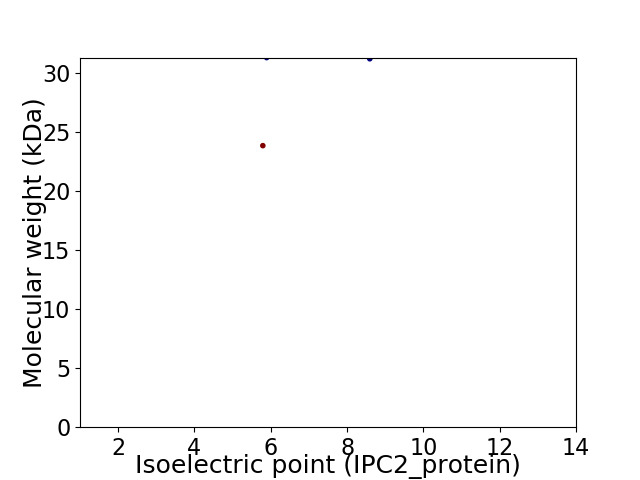
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
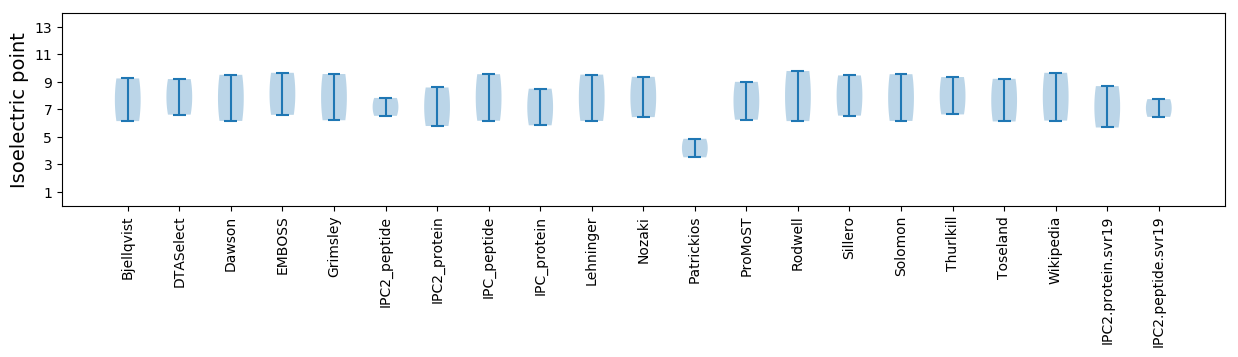
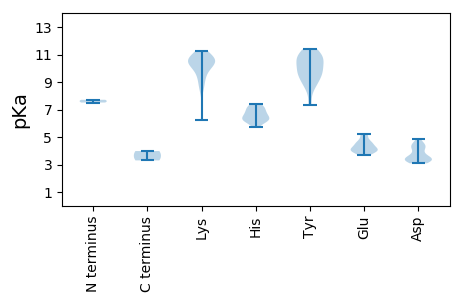
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RLS2|A0A0K1RLS2_9CIRC Putative replication initiation protein OS=Lytechinus variegatus variable sea urchin associated circular virus OX=1692254 PE=3 SV=1
MM1 pKa = 7.71PSFRR5 pKa = 11.84VCAKK9 pKa = 10.47NFFLTYY15 pKa = 9.45PRR17 pKa = 11.84SIDD20 pKa = 4.23LSKK23 pKa = 10.96QDD25 pKa = 4.83LFDD28 pKa = 4.34FLLSFEE34 pKa = 4.65PNYY37 pKa = 10.93LLVAEE42 pKa = 5.09EE43 pKa = 4.11KK44 pKa = 10.55HH45 pKa = 7.14ADD47 pKa = 3.83GTPHH51 pKa = 7.17LHH53 pKa = 7.38ALLCLARR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84DD63 pKa = 3.46IRR65 pKa = 11.84DD66 pKa = 3.3PRR68 pKa = 11.84HH69 pKa = 6.16FDD71 pKa = 3.33CQGYY75 pKa = 8.8HH76 pKa = 6.79CNITSTRR83 pKa = 11.84SIARR87 pKa = 11.84CIDD90 pKa = 3.24YY91 pKa = 10.59CKK93 pKa = 10.59KK94 pKa = 10.58DD95 pKa = 3.48DD96 pKa = 4.65CSPLSSGDD104 pKa = 3.38VPGRR108 pKa = 11.84DD109 pKa = 3.09GWGAICEE116 pKa = 4.35CGTRR120 pKa = 11.84EE121 pKa = 4.06DD122 pKa = 3.45FMGAVRR128 pKa = 11.84RR129 pKa = 11.84YY130 pKa = 8.68YY131 pKa = 10.14PRR133 pKa = 11.84DD134 pKa = 3.18YY135 pKa = 11.16VLNYY139 pKa = 9.54EE140 pKa = 4.52KK141 pKa = 11.02LLAYY145 pKa = 10.43ADD147 pKa = 3.37AHH149 pKa = 6.21FNANVPYY156 pKa = 9.78EE157 pKa = 4.0PQFTDD162 pKa = 4.31FVLPTEE168 pKa = 4.19VDD170 pKa = 3.48DD171 pKa = 4.39WLTDD175 pKa = 3.45NFKK178 pKa = 10.9VRR180 pKa = 11.84GEE182 pKa = 4.22NAFHH186 pKa = 6.4PTGGILIGPVVRR198 pKa = 11.84RR199 pKa = 11.84AFYY202 pKa = 9.44PLCKK206 pKa = 10.46GPP208 pKa = 3.98
MM1 pKa = 7.71PSFRR5 pKa = 11.84VCAKK9 pKa = 10.47NFFLTYY15 pKa = 9.45PRR17 pKa = 11.84SIDD20 pKa = 4.23LSKK23 pKa = 10.96QDD25 pKa = 4.83LFDD28 pKa = 4.34FLLSFEE34 pKa = 4.65PNYY37 pKa = 10.93LLVAEE42 pKa = 5.09EE43 pKa = 4.11KK44 pKa = 10.55HH45 pKa = 7.14ADD47 pKa = 3.83GTPHH51 pKa = 7.17LHH53 pKa = 7.38ALLCLARR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84DD63 pKa = 3.46IRR65 pKa = 11.84DD66 pKa = 3.3PRR68 pKa = 11.84HH69 pKa = 6.16FDD71 pKa = 3.33CQGYY75 pKa = 8.8HH76 pKa = 6.79CNITSTRR83 pKa = 11.84SIARR87 pKa = 11.84CIDD90 pKa = 3.24YY91 pKa = 10.59CKK93 pKa = 10.59KK94 pKa = 10.58DD95 pKa = 3.48DD96 pKa = 4.65CSPLSSGDD104 pKa = 3.38VPGRR108 pKa = 11.84DD109 pKa = 3.09GWGAICEE116 pKa = 4.35CGTRR120 pKa = 11.84EE121 pKa = 4.06DD122 pKa = 3.45FMGAVRR128 pKa = 11.84RR129 pKa = 11.84YY130 pKa = 8.68YY131 pKa = 10.14PRR133 pKa = 11.84DD134 pKa = 3.18YY135 pKa = 11.16VLNYY139 pKa = 9.54EE140 pKa = 4.52KK141 pKa = 11.02LLAYY145 pKa = 10.43ADD147 pKa = 3.37AHH149 pKa = 6.21FNANVPYY156 pKa = 9.78EE157 pKa = 4.0PQFTDD162 pKa = 4.31FVLPTEE168 pKa = 4.19VDD170 pKa = 3.48DD171 pKa = 4.39WLTDD175 pKa = 3.45NFKK178 pKa = 10.9VRR180 pKa = 11.84GEE182 pKa = 4.22NAFHH186 pKa = 6.4PTGGILIGPVVRR198 pKa = 11.84RR199 pKa = 11.84AFYY202 pKa = 9.44PLCKK206 pKa = 10.46GPP208 pKa = 3.98
Molecular weight: 23.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RLS2|A0A0K1RLS2_9CIRC Putative replication initiation protein OS=Lytechinus variegatus variable sea urchin associated circular virus OX=1692254 PE=3 SV=1
MM1 pKa = 7.51GNKK4 pKa = 9.42PLITNEE10 pKa = 3.72EE11 pKa = 4.39GQVSRR16 pKa = 11.84LYY18 pKa = 10.82APKK21 pKa = 10.24KK22 pKa = 6.22QTKK25 pKa = 9.36AGKK28 pKa = 9.9KK29 pKa = 8.58KK30 pKa = 9.69AEE32 pKa = 4.0RR33 pKa = 11.84YY34 pKa = 9.59KK35 pKa = 10.72RR36 pKa = 11.84FANKK40 pKa = 9.55VEE42 pKa = 4.09KK43 pKa = 10.64VINASLYY50 pKa = 8.86EE51 pKa = 3.74GRR53 pKa = 11.84XIHH56 pKa = 7.1RR57 pKa = 11.84SAIGSAVSSGDD68 pKa = 3.22AGKK71 pKa = 10.32QGLTSLMLFSAAGATNEE88 pKa = 4.65DD89 pKa = 4.49DD90 pKa = 3.56IEE92 pKa = 5.25KK93 pKa = 10.41IFAAEE98 pKa = 4.07GLTADD103 pKa = 4.27SDD105 pKa = 4.19EE106 pKa = 5.12IYY108 pKa = 10.91LKK110 pKa = 10.49SGYY113 pKa = 10.77LNATFKK119 pKa = 11.28CNTANVTGIVDD130 pKa = 4.0IYY132 pKa = 11.38QMEE135 pKa = 4.62SKK137 pKa = 10.63SAISVFNVKK146 pKa = 8.59DD147 pKa = 3.26TALPAFVTSVNDD159 pKa = 3.22QLAAQASATSTLTVTDD175 pKa = 4.49PGWTPFCNGSVGQHH189 pKa = 5.73FKK191 pKa = 10.84IKK193 pKa = 10.29DD194 pKa = 3.28HH195 pKa = 6.24TRR197 pKa = 11.84FICRR201 pKa = 11.84PGEE204 pKa = 3.97HH205 pKa = 6.19HH206 pKa = 6.72TFTYY210 pKa = 10.03RR211 pKa = 11.84KK212 pKa = 8.85KK213 pKa = 10.37ANRR216 pKa = 11.84SVKK219 pKa = 10.59LKK221 pKa = 10.46DD222 pKa = 3.61VKK224 pKa = 10.46EE225 pKa = 4.01LLMMPHH231 pKa = 6.52LTVGFLIILSGRR243 pKa = 11.84DD244 pKa = 3.51EE245 pKa = 4.59TVSTATVAGHH255 pKa = 6.3YY256 pKa = 7.34PTQTFDD262 pKa = 3.91FQWSRR267 pKa = 11.84TYY269 pKa = 9.83WYY271 pKa = 10.68NRR273 pKa = 11.84VHH275 pKa = 6.72ANVGFGGEE283 pKa = 3.89ISS285 pKa = 3.3
MM1 pKa = 7.51GNKK4 pKa = 9.42PLITNEE10 pKa = 3.72EE11 pKa = 4.39GQVSRR16 pKa = 11.84LYY18 pKa = 10.82APKK21 pKa = 10.24KK22 pKa = 6.22QTKK25 pKa = 9.36AGKK28 pKa = 9.9KK29 pKa = 8.58KK30 pKa = 9.69AEE32 pKa = 4.0RR33 pKa = 11.84YY34 pKa = 9.59KK35 pKa = 10.72RR36 pKa = 11.84FANKK40 pKa = 9.55VEE42 pKa = 4.09KK43 pKa = 10.64VINASLYY50 pKa = 8.86EE51 pKa = 3.74GRR53 pKa = 11.84XIHH56 pKa = 7.1RR57 pKa = 11.84SAIGSAVSSGDD68 pKa = 3.22AGKK71 pKa = 10.32QGLTSLMLFSAAGATNEE88 pKa = 4.65DD89 pKa = 4.49DD90 pKa = 3.56IEE92 pKa = 5.25KK93 pKa = 10.41IFAAEE98 pKa = 4.07GLTADD103 pKa = 4.27SDD105 pKa = 4.19EE106 pKa = 5.12IYY108 pKa = 10.91LKK110 pKa = 10.49SGYY113 pKa = 10.77LNATFKK119 pKa = 11.28CNTANVTGIVDD130 pKa = 4.0IYY132 pKa = 11.38QMEE135 pKa = 4.62SKK137 pKa = 10.63SAISVFNVKK146 pKa = 8.59DD147 pKa = 3.26TALPAFVTSVNDD159 pKa = 3.22QLAAQASATSTLTVTDD175 pKa = 4.49PGWTPFCNGSVGQHH189 pKa = 5.73FKK191 pKa = 10.84IKK193 pKa = 10.29DD194 pKa = 3.28HH195 pKa = 6.24TRR197 pKa = 11.84FICRR201 pKa = 11.84PGEE204 pKa = 3.97HH205 pKa = 6.19HH206 pKa = 6.72TFTYY210 pKa = 10.03RR211 pKa = 11.84KK212 pKa = 8.85KK213 pKa = 10.37ANRR216 pKa = 11.84SVKK219 pKa = 10.59LKK221 pKa = 10.46DD222 pKa = 3.61VKK224 pKa = 10.46EE225 pKa = 4.01LLMMPHH231 pKa = 6.52LTVGFLIILSGRR243 pKa = 11.84DD244 pKa = 3.51EE245 pKa = 4.59TVSTATVAGHH255 pKa = 6.3YY256 pKa = 7.34PTQTFDD262 pKa = 3.91FQWSRR267 pKa = 11.84TYY269 pKa = 9.83WYY271 pKa = 10.68NRR273 pKa = 11.84VHH275 pKa = 6.72ANVGFGGEE283 pKa = 3.89ISS285 pKa = 3.3
Molecular weight: 31.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
493 |
208 |
285 |
246.5 |
27.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.316 ± 0.941 | 2.637 ± 1.289 |
6.491 ± 1.57 | 4.665 ± 0.201 |
5.68 ± 0.624 | 7.099 ± 0.504 |
3.043 ± 0.192 | 4.665 ± 0.486 |
6.288 ± 1.45 | 7.505 ± 0.967 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.42 ± 0.272 | 4.462 ± 0.366 |
4.665 ± 1.512 | 2.434 ± 0.589 |
5.882 ± 1.36 | 6.288 ± 1.164 |
6.897 ± 1.526 | 6.085 ± 0.473 |
1.014 ± 0.031 | 4.26 ± 0.611 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
