
Mycobacterium phage Henu3 PeY-2017
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Cheoctovirus; unclassified Cheoctovirus
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
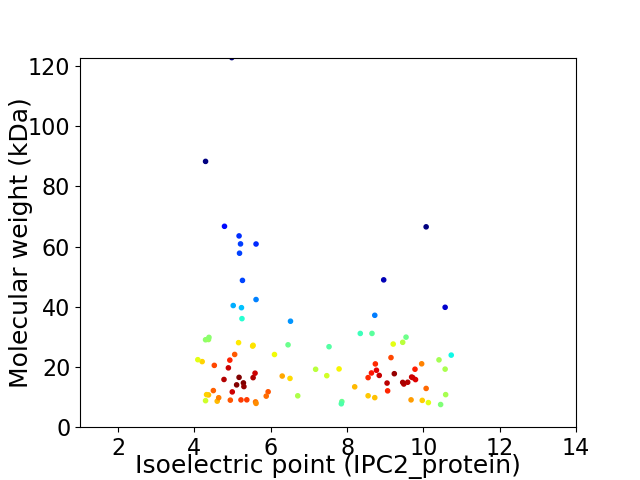
Virtual 2D-PAGE plot for 95 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
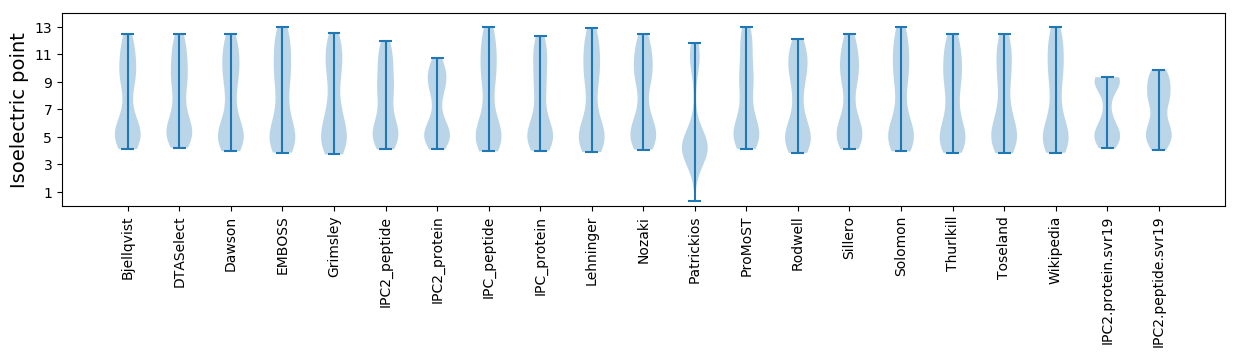
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A481W3V7|A0A481W3V7_9CAUD Portal protein OS=Mycobacterium phage Henu3 PeY-2017 OX=2571241 GN=Henu3_gp1 PE=4 SV=1
MM1 pKa = 7.82GEE3 pKa = 4.37PEE5 pKa = 5.42DD6 pKa = 3.94PTEE9 pKa = 4.09LAGVADD15 pKa = 5.09ADD17 pKa = 4.13TMSAYY22 pKa = 9.87AWSVEE27 pKa = 3.79DD28 pKa = 3.46PTINSEE34 pKa = 4.8LIDD37 pKa = 4.21AGSDD41 pKa = 3.22RR42 pKa = 11.84PFWITTAAVGLSLSLVAVVGVLGYY66 pKa = 9.98RR67 pKa = 11.84VWVDD71 pKa = 3.29VPEE74 pKa = 4.65RR75 pKa = 11.84PTVVVAATPTTTTEE89 pKa = 3.95VEE91 pKa = 4.2AASPLPLPPPVTVTTVVVQTPAPAPAPVPVPEE123 pKa = 4.97VIPSVLPTPLPPLSNTDD140 pKa = 3.3MVFLSRR146 pKa = 11.84MRR148 pKa = 11.84AQGWYY153 pKa = 10.37VSDD156 pKa = 4.2PQLMAYY162 pKa = 10.16RR163 pKa = 11.84AFEE166 pKa = 4.09TCAMLRR172 pKa = 11.84NGEE175 pKa = 4.45PVWQVQGKK183 pKa = 9.02LLGLEE188 pKa = 4.23GVPNEE193 pKa = 4.14VEE195 pKa = 3.55ASKK198 pKa = 10.7FLNTAMSTYY207 pKa = 10.04PNCPP211 pKa = 3.13
MM1 pKa = 7.82GEE3 pKa = 4.37PEE5 pKa = 5.42DD6 pKa = 3.94PTEE9 pKa = 4.09LAGVADD15 pKa = 5.09ADD17 pKa = 4.13TMSAYY22 pKa = 9.87AWSVEE27 pKa = 3.79DD28 pKa = 3.46PTINSEE34 pKa = 4.8LIDD37 pKa = 4.21AGSDD41 pKa = 3.22RR42 pKa = 11.84PFWITTAAVGLSLSLVAVVGVLGYY66 pKa = 9.98RR67 pKa = 11.84VWVDD71 pKa = 3.29VPEE74 pKa = 4.65RR75 pKa = 11.84PTVVVAATPTTTTEE89 pKa = 3.95VEE91 pKa = 4.2AASPLPLPPPVTVTTVVVQTPAPAPAPVPVPEE123 pKa = 4.97VIPSVLPTPLPPLSNTDD140 pKa = 3.3MVFLSRR146 pKa = 11.84MRR148 pKa = 11.84AQGWYY153 pKa = 10.37VSDD156 pKa = 4.2PQLMAYY162 pKa = 10.16RR163 pKa = 11.84AFEE166 pKa = 4.09TCAMLRR172 pKa = 11.84NGEE175 pKa = 4.45PVWQVQGKK183 pKa = 9.02LLGLEE188 pKa = 4.23GVPNEE193 pKa = 4.14VEE195 pKa = 3.55ASKK198 pKa = 10.7FLNTAMSTYY207 pKa = 10.04PNCPP211 pKa = 3.13
Molecular weight: 22.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A481W3X6|A0A481W3X6_9CAUD Uncharacterized protein OS=Mycobacterium phage Henu3 PeY-2017 OX=2571241 GN=Henu3_gp43 PE=4 SV=1
MM1 pKa = 7.31LVLRR5 pKa = 11.84RR6 pKa = 11.84GSPRR10 pKa = 11.84GTASIKK16 pKa = 10.61PPTPSRR22 pKa = 11.84NAPSPTPTSIGRR34 pKa = 11.84SANSKK39 pKa = 7.62QTSTPTSPTSRR50 pKa = 11.84QRR52 pKa = 11.84SVSALRR58 pKa = 11.84NCSNTSRR65 pKa = 11.84KK66 pKa = 9.69NDD68 pKa = 3.58GNRR71 pKa = 11.84KK72 pKa = 8.17RR73 pKa = 11.84CSGRR77 pKa = 11.84HH78 pKa = 4.13STSDD82 pKa = 2.86QADD85 pKa = 3.66RR86 pKa = 11.84HH87 pKa = 5.66RR88 pKa = 11.84RR89 pKa = 11.84PCGNTHH95 pKa = 5.8KK96 pKa = 9.03TQTHH100 pKa = 6.45EE101 pKa = 4.07PQQTPPHH108 pKa = 6.98PLQAHH113 pKa = 6.32HH114 pKa = 7.02RR115 pKa = 11.84LAPPSHH121 pKa = 6.66PTPHH125 pKa = 6.09TTTPTNPPTHH135 pKa = 6.26QNPHH139 pKa = 5.64LRR141 pKa = 11.84TPRR144 pKa = 11.84RR145 pKa = 11.84MGQRR149 pKa = 11.84HH150 pKa = 5.82RR151 pKa = 11.84STHH154 pKa = 5.4RR155 pKa = 11.84RR156 pKa = 11.84HH157 pKa = 4.89AHH159 pKa = 5.43KK160 pKa = 10.32LARR163 pKa = 11.84LPRR166 pKa = 11.84RR167 pKa = 11.84TTQRR171 pKa = 11.84NPTTPRR177 pKa = 11.84KK178 pKa = 8.54RR179 pKa = 11.84TKK181 pKa = 8.77TNHH184 pKa = 5.71RR185 pKa = 11.84RR186 pKa = 11.84LEE188 pKa = 4.01IPRR191 pKa = 11.84TTLRR195 pKa = 11.84TTHH198 pKa = 6.22PTRR201 pKa = 11.84HH202 pKa = 6.01PRR204 pKa = 11.84RR205 pKa = 11.84PQRR208 pKa = 11.84TTT210 pKa = 2.68
MM1 pKa = 7.31LVLRR5 pKa = 11.84RR6 pKa = 11.84GSPRR10 pKa = 11.84GTASIKK16 pKa = 10.61PPTPSRR22 pKa = 11.84NAPSPTPTSIGRR34 pKa = 11.84SANSKK39 pKa = 7.62QTSTPTSPTSRR50 pKa = 11.84QRR52 pKa = 11.84SVSALRR58 pKa = 11.84NCSNTSRR65 pKa = 11.84KK66 pKa = 9.69NDD68 pKa = 3.58GNRR71 pKa = 11.84KK72 pKa = 8.17RR73 pKa = 11.84CSGRR77 pKa = 11.84HH78 pKa = 4.13STSDD82 pKa = 2.86QADD85 pKa = 3.66RR86 pKa = 11.84HH87 pKa = 5.66RR88 pKa = 11.84RR89 pKa = 11.84PCGNTHH95 pKa = 5.8KK96 pKa = 9.03TQTHH100 pKa = 6.45EE101 pKa = 4.07PQQTPPHH108 pKa = 6.98PLQAHH113 pKa = 6.32HH114 pKa = 7.02RR115 pKa = 11.84LAPPSHH121 pKa = 6.66PTPHH125 pKa = 6.09TTTPTNPPTHH135 pKa = 6.26QNPHH139 pKa = 5.64LRR141 pKa = 11.84TPRR144 pKa = 11.84RR145 pKa = 11.84MGQRR149 pKa = 11.84HH150 pKa = 5.82RR151 pKa = 11.84STHH154 pKa = 5.4RR155 pKa = 11.84RR156 pKa = 11.84HH157 pKa = 4.89AHH159 pKa = 5.43KK160 pKa = 10.32LARR163 pKa = 11.84LPRR166 pKa = 11.84RR167 pKa = 11.84TTQRR171 pKa = 11.84NPTTPRR177 pKa = 11.84KK178 pKa = 8.54RR179 pKa = 11.84TKK181 pKa = 8.77TNHH184 pKa = 5.71RR185 pKa = 11.84RR186 pKa = 11.84LEE188 pKa = 4.01IPRR191 pKa = 11.84TTLRR195 pKa = 11.84TTHH198 pKa = 6.22PTRR201 pKa = 11.84HH202 pKa = 6.01PRR204 pKa = 11.84RR205 pKa = 11.84PQRR208 pKa = 11.84TTT210 pKa = 2.68
Molecular weight: 23.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
20957 |
69 |
1224 |
220.6 |
24.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.791 ± 0.464 | 1.546 ± 0.182 |
5.602 ± 0.181 | 4.853 ± 0.271 |
3.011 ± 0.21 | 8.551 ± 0.5 |
2.987 ± 0.273 | 3.937 ± 0.227 |
2.729 ± 0.18 | 7.616 ± 0.206 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.104 ± 0.124 | 3.483 ± 0.156 |
6.886 ± 0.237 | 3.588 ± 0.187 |
8.37 ± 0.549 | 6.847 ± 0.342 |
6.814 ± 0.268 | 7.191 ± 0.298 |
1.999 ± 0.162 | 2.095 ± 0.152 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
