
Beihai sphaeromadae virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
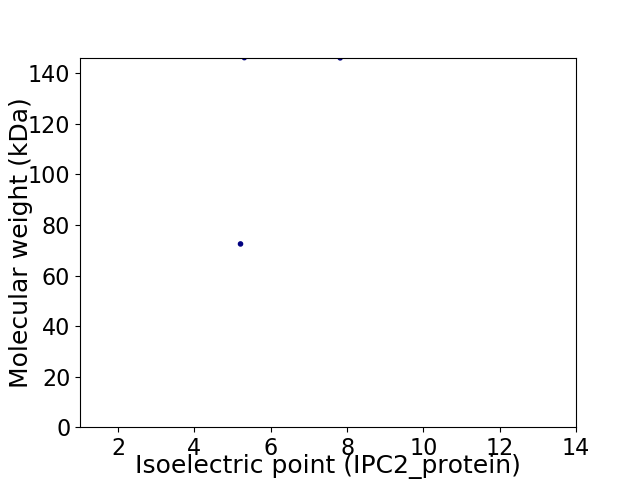
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHV5|A0A1L3KHV5_9VIRU Putative capsid protein OS=Beihai sphaeromadae virus 4 OX=1922710 PE=4 SV=1
MM1 pKa = 7.68PSRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84IRR8 pKa = 11.84QPIRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84MPKK17 pKa = 10.11RR18 pKa = 11.84IRR20 pKa = 11.84QPKK23 pKa = 9.08PVTSSQTLTSTYY35 pKa = 9.88DD36 pKa = 3.18IPIGLPSMTAFSTKK50 pKa = 10.14AVSLDD55 pKa = 3.47PSAHH59 pKa = 5.58EE60 pKa = 4.15NSRR63 pKa = 11.84ASAQMSIYY71 pKa = 10.86NKK73 pKa = 10.25YY74 pKa = 10.37KK75 pKa = 10.83LLACSLIYY83 pKa = 10.51SPSVGYY89 pKa = 10.49DD90 pKa = 3.04SSSGLLGVFHH100 pKa = 6.55STAEE104 pKa = 4.07EE105 pKa = 4.16EE106 pKa = 4.32IAPQTRR112 pKa = 11.84EE113 pKa = 4.0DD114 pKa = 3.64FQGRR118 pKa = 11.84IQNAGPSGCLIPVRR132 pKa = 11.84KK133 pKa = 9.84NKK135 pKa = 9.64VFTVPKK141 pKa = 10.21SVIRR145 pKa = 11.84NIAEE149 pKa = 4.67GYY151 pKa = 8.26TPGATQTPGRR161 pKa = 11.84IVIGSATSCDD171 pKa = 3.19IDD173 pKa = 4.45SPGLLTLRR181 pKa = 11.84STYY184 pKa = 10.15QFIGPTVPATPASPTKK200 pKa = 9.94IIKK203 pKa = 9.45RR204 pKa = 11.84APYY207 pKa = 8.69TRR209 pKa = 11.84RR210 pKa = 11.84LASFVWKK217 pKa = 7.95PTNPEE222 pKa = 3.36WFLISGLVDD231 pKa = 3.62SYY233 pKa = 11.89AATSDD238 pKa = 4.02AITDD242 pKa = 3.35VSIEE246 pKa = 3.82FSVYY250 pKa = 9.45NASGSSFGKK259 pKa = 10.59SIGQSYY265 pKa = 11.16NDD267 pKa = 3.37WQSFIEE273 pKa = 4.4SAFADD278 pKa = 3.92SQADD282 pKa = 3.45WEE284 pKa = 4.81TFTSSTLAAITFDD297 pKa = 3.98PPGSPFKK304 pKa = 10.37TAGILDD310 pKa = 3.73SQRR313 pKa = 11.84FVTTMPDD320 pKa = 2.99STTTTLVDD328 pKa = 3.65SVTTTLEE335 pKa = 3.92EE336 pKa = 4.58SVTTTLPLSQTTTLQDD352 pKa = 3.32SVTTTLQEE360 pKa = 4.44SVTTTLPEE368 pKa = 4.27SQTTTLEE375 pKa = 4.1DD376 pKa = 3.69SKK378 pKa = 9.02TTTLDD383 pKa = 3.33SSVTTTLPQSQTTTLQGSIVKK404 pKa = 8.58TEE406 pKa = 4.33PGSVTTTDD414 pKa = 3.16SDD416 pKa = 4.08STTKK420 pKa = 9.99TLSGSVTTTDD430 pKa = 3.71EE431 pKa = 3.99NSKK434 pKa = 7.89TTTQIEE440 pKa = 4.4SRR442 pKa = 11.84VYY444 pKa = 10.02IALPNQGVVSSNNPLPVDD462 pKa = 4.04DD463 pKa = 4.91GGGGFFMNLATGVITAFVLEE483 pKa = 4.56KK484 pKa = 10.28GRR486 pKa = 11.84KK487 pKa = 8.12YY488 pKa = 11.03GPDD491 pKa = 3.13SKK493 pKa = 9.86EE494 pKa = 4.01TPEE497 pKa = 4.53GYY499 pKa = 10.3VRR501 pKa = 11.84LKK503 pKa = 9.53GTYY506 pKa = 8.91PAGGDD511 pKa = 3.44VDD513 pKa = 4.19GVSYY517 pKa = 10.92PDD519 pKa = 3.4VNAPSSTTSVDD530 pKa = 3.38YY531 pKa = 11.09KK532 pKa = 10.82QFTSFDD538 pKa = 3.81VLIKK542 pKa = 9.79PQNFEE547 pKa = 3.79GTGWHH552 pKa = 5.85QKK554 pKa = 10.1LDD556 pKa = 3.91SVICRR561 pKa = 11.84HH562 pKa = 5.5YY563 pKa = 10.7LGPAYY568 pKa = 10.29YY569 pKa = 10.91GNLQSRR575 pKa = 11.84VAIDD579 pKa = 3.4VTVAPILFEE588 pKa = 4.54EE589 pKa = 5.17ADD591 pKa = 3.72TGNKK595 pKa = 9.38ILGDD599 pKa = 3.62PCRR602 pKa = 11.84YY603 pKa = 9.13RR604 pKa = 11.84ALYY607 pKa = 10.31YY608 pKa = 10.59KK609 pKa = 9.45DD610 pKa = 3.69TNDD613 pKa = 2.94FRR615 pKa = 11.84VIRR618 pKa = 11.84SSTEE622 pKa = 3.79VEE624 pKa = 3.69AKK626 pKa = 10.66LYY628 pKa = 7.68DD629 pKa = 3.66TQSAIGFRR637 pKa = 11.84VIPVMSDD644 pKa = 3.07YY645 pKa = 10.86PIYY648 pKa = 10.51EE649 pKa = 4.79DD650 pKa = 5.12EE651 pKa = 4.45SKK653 pKa = 8.84TWFAIYY659 pKa = 9.87GHH661 pKa = 6.24NVDD664 pKa = 4.14TNLL667 pKa = 4.15
MM1 pKa = 7.68PSRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84IRR8 pKa = 11.84QPIRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84MPKK17 pKa = 10.11RR18 pKa = 11.84IRR20 pKa = 11.84QPKK23 pKa = 9.08PVTSSQTLTSTYY35 pKa = 9.88DD36 pKa = 3.18IPIGLPSMTAFSTKK50 pKa = 10.14AVSLDD55 pKa = 3.47PSAHH59 pKa = 5.58EE60 pKa = 4.15NSRR63 pKa = 11.84ASAQMSIYY71 pKa = 10.86NKK73 pKa = 10.25YY74 pKa = 10.37KK75 pKa = 10.83LLACSLIYY83 pKa = 10.51SPSVGYY89 pKa = 10.49DD90 pKa = 3.04SSSGLLGVFHH100 pKa = 6.55STAEE104 pKa = 4.07EE105 pKa = 4.16EE106 pKa = 4.32IAPQTRR112 pKa = 11.84EE113 pKa = 4.0DD114 pKa = 3.64FQGRR118 pKa = 11.84IQNAGPSGCLIPVRR132 pKa = 11.84KK133 pKa = 9.84NKK135 pKa = 9.64VFTVPKK141 pKa = 10.21SVIRR145 pKa = 11.84NIAEE149 pKa = 4.67GYY151 pKa = 8.26TPGATQTPGRR161 pKa = 11.84IVIGSATSCDD171 pKa = 3.19IDD173 pKa = 4.45SPGLLTLRR181 pKa = 11.84STYY184 pKa = 10.15QFIGPTVPATPASPTKK200 pKa = 9.94IIKK203 pKa = 9.45RR204 pKa = 11.84APYY207 pKa = 8.69TRR209 pKa = 11.84RR210 pKa = 11.84LASFVWKK217 pKa = 7.95PTNPEE222 pKa = 3.36WFLISGLVDD231 pKa = 3.62SYY233 pKa = 11.89AATSDD238 pKa = 4.02AITDD242 pKa = 3.35VSIEE246 pKa = 3.82FSVYY250 pKa = 9.45NASGSSFGKK259 pKa = 10.59SIGQSYY265 pKa = 11.16NDD267 pKa = 3.37WQSFIEE273 pKa = 4.4SAFADD278 pKa = 3.92SQADD282 pKa = 3.45WEE284 pKa = 4.81TFTSSTLAAITFDD297 pKa = 3.98PPGSPFKK304 pKa = 10.37TAGILDD310 pKa = 3.73SQRR313 pKa = 11.84FVTTMPDD320 pKa = 2.99STTTTLVDD328 pKa = 3.65SVTTTLEE335 pKa = 3.92EE336 pKa = 4.58SVTTTLPLSQTTTLQDD352 pKa = 3.32SVTTTLQEE360 pKa = 4.44SVTTTLPEE368 pKa = 4.27SQTTTLEE375 pKa = 4.1DD376 pKa = 3.69SKK378 pKa = 9.02TTTLDD383 pKa = 3.33SSVTTTLPQSQTTTLQGSIVKK404 pKa = 8.58TEE406 pKa = 4.33PGSVTTTDD414 pKa = 3.16SDD416 pKa = 4.08STTKK420 pKa = 9.99TLSGSVTTTDD430 pKa = 3.71EE431 pKa = 3.99NSKK434 pKa = 7.89TTTQIEE440 pKa = 4.4SRR442 pKa = 11.84VYY444 pKa = 10.02IALPNQGVVSSNNPLPVDD462 pKa = 4.04DD463 pKa = 4.91GGGGFFMNLATGVITAFVLEE483 pKa = 4.56KK484 pKa = 10.28GRR486 pKa = 11.84KK487 pKa = 8.12YY488 pKa = 11.03GPDD491 pKa = 3.13SKK493 pKa = 9.86EE494 pKa = 4.01TPEE497 pKa = 4.53GYY499 pKa = 10.3VRR501 pKa = 11.84LKK503 pKa = 9.53GTYY506 pKa = 8.91PAGGDD511 pKa = 3.44VDD513 pKa = 4.19GVSYY517 pKa = 10.92PDD519 pKa = 3.4VNAPSSTTSVDD530 pKa = 3.38YY531 pKa = 11.09KK532 pKa = 10.82QFTSFDD538 pKa = 3.81VLIKK542 pKa = 9.79PQNFEE547 pKa = 3.79GTGWHH552 pKa = 5.85QKK554 pKa = 10.1LDD556 pKa = 3.91SVICRR561 pKa = 11.84HH562 pKa = 5.5YY563 pKa = 10.7LGPAYY568 pKa = 10.29YY569 pKa = 10.91GNLQSRR575 pKa = 11.84VAIDD579 pKa = 3.4VTVAPILFEE588 pKa = 4.54EE589 pKa = 5.17ADD591 pKa = 3.72TGNKK595 pKa = 9.38ILGDD599 pKa = 3.62PCRR602 pKa = 11.84YY603 pKa = 9.13RR604 pKa = 11.84ALYY607 pKa = 10.31YY608 pKa = 10.59KK609 pKa = 9.45DD610 pKa = 3.69TNDD613 pKa = 2.94FRR615 pKa = 11.84VIRR618 pKa = 11.84SSTEE622 pKa = 3.79VEE624 pKa = 3.69AKK626 pKa = 10.66LYY628 pKa = 7.68DD629 pKa = 3.66TQSAIGFRR637 pKa = 11.84VIPVMSDD644 pKa = 3.07YY645 pKa = 10.86PIYY648 pKa = 10.51EE649 pKa = 4.79DD650 pKa = 5.12EE651 pKa = 4.45SKK653 pKa = 8.84TWFAIYY659 pKa = 9.87GHH661 pKa = 6.24NVDD664 pKa = 4.14TNLL667 pKa = 4.15
Molecular weight: 72.51 kDa
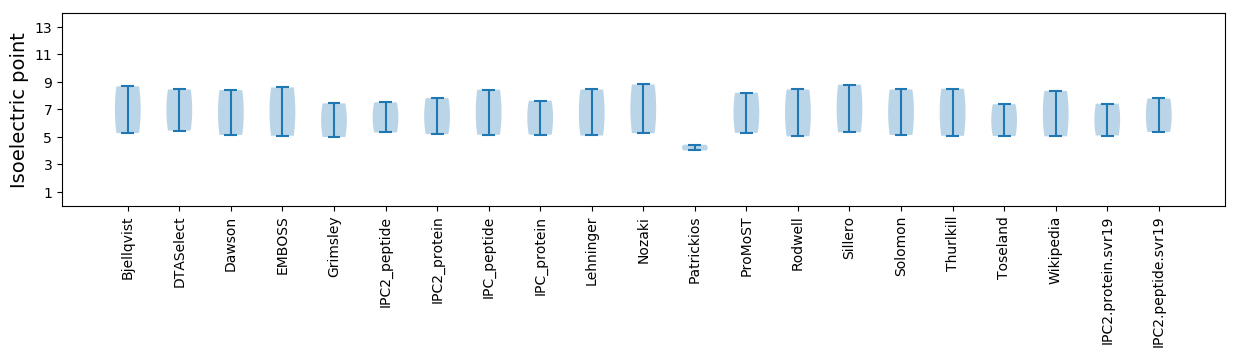
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHV5|A0A1L3KHV5_9VIRU Putative capsid protein OS=Beihai sphaeromadae virus 4 OX=1922710 PE=4 SV=1
MM1 pKa = 7.76ASFSNPVNASEE12 pKa = 4.43KK13 pKa = 10.62LSGASFLAVATRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84QDD30 pKa = 3.05LSFLEE35 pKa = 4.52GQLDD39 pKa = 3.38KK40 pKa = 10.59CTRR43 pKa = 11.84LSPQFHH49 pKa = 6.55LPIRR53 pKa = 11.84DD54 pKa = 3.24IPEE57 pKa = 3.87IKK59 pKa = 9.44STGKK63 pKa = 11.03GDD65 pKa = 3.41MQRR68 pKa = 11.84AKK70 pKa = 9.41TQTFDD75 pKa = 3.82VPDD78 pKa = 4.05NFEE81 pKa = 4.59KK82 pKa = 10.88LPPIPNSKK90 pKa = 8.61LTRR93 pKa = 11.84NGEE96 pKa = 4.3LIQKK100 pKa = 9.97GNLGKK105 pKa = 10.37RR106 pKa = 11.84FYY108 pKa = 11.09NSQKK112 pKa = 9.23TFGGKK117 pKa = 8.8NVPDD121 pKa = 4.13NLRR124 pKa = 11.84QTIRR128 pKa = 11.84HH129 pKa = 5.59ALATRR134 pKa = 11.84IEE136 pKa = 4.38INGPAVEE143 pKa = 4.39SCVIADD149 pKa = 3.71EE150 pKa = 4.78LMATGLEE157 pKa = 4.2WEE159 pKa = 4.59YY160 pKa = 11.59SAGTTQGLVEE170 pKa = 4.02RR171 pKa = 11.84LSRR174 pKa = 11.84FGKK177 pKa = 10.05DD178 pKa = 2.92DD179 pKa = 3.29KK180 pKa = 10.83TVSFFRR186 pKa = 11.84QTDD189 pKa = 3.91EE190 pKa = 3.57IRR192 pKa = 11.84NFQSFLFQALDD203 pKa = 3.28TLLPLKK209 pKa = 9.03STPFEE214 pKa = 3.91TSFPSTSTEE223 pKa = 3.9VIEE226 pKa = 4.97GIRR229 pKa = 11.84VNPQSSAGPMYY240 pKa = 9.96GTTKK244 pKa = 10.39VQALDD249 pKa = 5.17DD250 pKa = 3.73IMLTMDD256 pKa = 5.59AIRR259 pKa = 11.84TQFSLGTLDD268 pKa = 3.53TFFAAHH274 pKa = 7.51PEE276 pKa = 4.04TLIADD281 pKa = 4.28CRR283 pKa = 11.84NKK285 pKa = 9.38TDD287 pKa = 3.18RR288 pKa = 11.84YY289 pKa = 9.91EE290 pKa = 3.98IAKK293 pKa = 10.14LPEE296 pKa = 3.66KK297 pKa = 9.66TRR299 pKa = 11.84PYY301 pKa = 10.33WSFNAHH307 pKa = 3.89WQFYY311 pKa = 8.49YY312 pKa = 10.41SWLMQRR318 pKa = 11.84FCDD321 pKa = 3.84NLYY324 pKa = 9.98VASDD328 pKa = 4.16PEE330 pKa = 4.95CDD332 pKa = 3.31VPTYY336 pKa = 10.94NYY338 pKa = 9.99YY339 pKa = 10.07GHH341 pKa = 6.53VWAKK345 pKa = 10.64GGAQRR350 pKa = 11.84IHH352 pKa = 6.17EE353 pKa = 4.83SIMNVAKK360 pKa = 10.01TGRR363 pKa = 11.84PFVGIYY369 pKa = 10.82GDD371 pKa = 4.08DD372 pKa = 3.63FLFAFPIKK380 pKa = 9.4TDD382 pKa = 3.25EE383 pKa = 4.27GTTIQVMAPDD393 pKa = 4.43ISQMDD398 pKa = 3.94STVSHH403 pKa = 5.93AVIRR407 pKa = 11.84GTCDD411 pKa = 3.23YY412 pKa = 10.54IYY414 pKa = 10.71KK415 pKa = 10.12QYY417 pKa = 11.08QEE419 pKa = 4.29AHH421 pKa = 6.44GDD423 pKa = 3.65DD424 pKa = 3.76PFVRR428 pKa = 11.84FLLTRR433 pKa = 11.84LAEE436 pKa = 4.57DD437 pKa = 3.34ASSPRR442 pKa = 11.84FLASAIATYY451 pKa = 10.49QNQRR455 pKa = 11.84PTGLATGVVGTTLFDD470 pKa = 3.64TVQAALLAFSVIEE483 pKa = 3.68EE484 pKa = 4.11WNRR487 pKa = 11.84TKK489 pKa = 10.84RR490 pKa = 11.84SPDD493 pKa = 3.23EE494 pKa = 3.99IFNKK498 pKa = 9.52VSKK501 pKa = 10.43KK502 pKa = 10.02FGMRR506 pKa = 11.84AKK508 pKa = 10.42SLEE511 pKa = 3.94LYY513 pKa = 9.45EE514 pKa = 4.55YY515 pKa = 10.94NGLPPAEE522 pKa = 4.08GTEE525 pKa = 4.08WTFLGMQLATYY536 pKa = 10.49SNSKK540 pKa = 7.91HH541 pKa = 5.35TNGVIPRR548 pKa = 11.84LPRR551 pKa = 11.84DD552 pKa = 3.56SLLKK556 pKa = 10.72SILNPRR562 pKa = 11.84INSYY566 pKa = 10.77LFSSKK571 pKa = 10.64LGTQRR576 pKa = 11.84YY577 pKa = 8.91LFDD580 pKa = 5.0ASRR583 pKa = 11.84GWYY586 pKa = 9.86LCALGDD592 pKa = 3.78RR593 pKa = 11.84LIWDD597 pKa = 3.73VLSSVIEE604 pKa = 4.27TVPTMAVGMVVQAGGGLGAPPEE626 pKa = 4.46CNGLFDD632 pKa = 4.79VNRR635 pKa = 11.84PFNWLSSFGVPTMDD649 pKa = 2.77WVADD653 pKa = 4.05LFLSPMDD660 pKa = 4.97PDD662 pKa = 3.78PAGWLSLLDD671 pKa = 3.86PPKK674 pKa = 10.78GRR676 pKa = 11.84LIVEE680 pKa = 4.29MQDD683 pKa = 3.26QVDD686 pKa = 4.11LSRR689 pKa = 11.84YY690 pKa = 6.89TIRR693 pKa = 11.84PTPNTAPHH701 pKa = 5.44SWEE704 pKa = 3.81TRR706 pKa = 11.84EE707 pKa = 4.99KK708 pKa = 10.13VTDD711 pKa = 3.62TNLEE715 pKa = 4.19SLVSDD720 pKa = 3.88NPSGLGPQVQRR731 pKa = 11.84PKK733 pKa = 10.51AQHH736 pKa = 6.23LPPVKK741 pKa = 10.41APPFKK746 pKa = 10.41IRR748 pKa = 11.84LSEE751 pKa = 4.1HH752 pKa = 6.27LSKK755 pKa = 11.18HH756 pKa = 5.96NITAFVPSEE765 pKa = 3.76LAAEE769 pKa = 4.53LGVSVTTILTEE780 pKa = 4.41LYY782 pKa = 10.45SKK784 pKa = 10.52EE785 pKa = 4.33RR786 pKa = 11.84EE787 pKa = 4.06TKK789 pKa = 9.72APEE792 pKa = 3.78FRR794 pKa = 11.84WYY796 pKa = 10.51SLGRR800 pKa = 11.84ILTKK804 pKa = 10.9DD805 pKa = 3.29PVDD808 pKa = 3.55TVQPSRR814 pKa = 11.84ILWNQYY820 pKa = 9.05SRR822 pKa = 11.84ILSEE826 pKa = 4.63LGYY829 pKa = 10.34SNPAIQPARR838 pKa = 11.84PKK840 pKa = 10.27AGPVPAPRR848 pKa = 11.84YY849 pKa = 10.25SKK851 pKa = 10.26MKK853 pKa = 10.78ANVPQDD859 pKa = 3.47FATSNSTTISDD870 pKa = 4.63CDD872 pKa = 3.4TSPAIRR878 pKa = 11.84VSGPDD883 pKa = 3.23QTPTSQTVPQYY894 pKa = 9.79TPKK897 pKa = 9.26PAQMMYY903 pKa = 10.64QRR905 pKa = 11.84IPAKK909 pKa = 9.83TGAVVCALEE918 pKa = 4.36EE919 pKa = 4.08NLVAPRR925 pKa = 11.84AEE927 pKa = 4.3GLSSGARR934 pKa = 11.84VPLADD939 pKa = 3.46QVNSGRR945 pKa = 11.84IVHH948 pKa = 6.36QYY950 pKa = 10.35FNWDD954 pKa = 3.57PPSPILSTSPEE965 pKa = 3.76QTLPLDD971 pKa = 4.23LSTSEE976 pKa = 4.65SVSTPDD982 pKa = 3.19TTSTPDD988 pKa = 2.96TVMYY992 pKa = 9.92RR993 pKa = 11.84SPPIEE998 pKa = 4.25GSQCYY1003 pKa = 9.95EE1004 pKa = 3.96EE1005 pKa = 4.8DD1006 pKa = 3.38QSDD1009 pKa = 3.84GSEE1012 pKa = 4.29SSWTSYY1018 pKa = 10.88ASSVSGCQRR1027 pKa = 11.84FDD1029 pKa = 3.37YY1030 pKa = 10.32PNRR1033 pKa = 11.84RR1034 pKa = 11.84SHH1036 pKa = 7.51PDD1038 pKa = 2.9AFPNQEE1044 pKa = 3.94AQSFRR1049 pKa = 11.84AAKK1052 pKa = 8.96EE1053 pKa = 3.46QYY1055 pKa = 9.72EE1056 pKa = 4.56RR1057 pKa = 11.84YY1058 pKa = 7.56TEE1060 pKa = 4.01SRR1062 pKa = 11.84RR1063 pKa = 11.84TQSPPQSPPPRR1074 pKa = 11.84WPTPPPPIPITEE1086 pKa = 4.3GSLTYY1091 pKa = 10.23TSVSTTTSNSKK1102 pKa = 9.97QVASGDD1108 pKa = 3.47RR1109 pKa = 11.84SRR1111 pKa = 11.84GSGAKK1116 pKa = 9.59PKK1118 pKa = 10.68VSTHH1122 pKa = 5.33SKK1124 pKa = 10.43KK1125 pKa = 9.9STDD1128 pKa = 3.36VPSQVPVKK1136 pKa = 10.2SVEE1139 pKa = 4.1VKK1141 pKa = 10.59GRR1143 pKa = 11.84TFTNSTRR1150 pKa = 11.84FSGVEE1155 pKa = 3.79HH1156 pKa = 6.8KK1157 pKa = 10.07PVKK1160 pKa = 7.75TTNGNQRR1167 pKa = 11.84VFYY1170 pKa = 10.0KK1171 pKa = 10.82VKK1173 pKa = 10.69SKK1175 pKa = 10.94DD1176 pKa = 3.37FRR1178 pKa = 11.84HH1179 pKa = 6.24PPVPPTIVGFYY1190 pKa = 10.21DD1191 pKa = 3.98GKK1193 pKa = 10.22HH1194 pKa = 5.39NYY1196 pKa = 8.57YY1197 pKa = 10.44VPTYY1201 pKa = 9.69KK1202 pKa = 10.61CQVDD1206 pKa = 3.95SKK1208 pKa = 9.22VKK1210 pKa = 10.5CVNTPPNPTKK1220 pKa = 10.62LGNFAWLGSGVSSLCAWAVTGKK1242 pKa = 10.64FEE1244 pKa = 5.13LVFDD1248 pKa = 4.95FTHH1251 pKa = 6.51QPDD1254 pKa = 3.66KK1255 pKa = 11.05MLFQCSIYY1263 pKa = 10.06IDD1265 pKa = 3.87GKK1267 pKa = 11.32LMISASRR1274 pKa = 11.84CKK1276 pKa = 10.2SKK1278 pKa = 11.16KK1279 pKa = 7.04EE1280 pKa = 3.95AKK1282 pKa = 9.91EE1283 pKa = 3.3IAAFQFLHH1291 pKa = 6.15SCFLHH1296 pKa = 5.22RR1297 pKa = 11.84TPRR1300 pKa = 11.84DD1301 pKa = 3.88LIHH1304 pKa = 6.85HH1305 pKa = 6.67ALKK1308 pKa = 9.84TPHH1311 pKa = 6.49
MM1 pKa = 7.76ASFSNPVNASEE12 pKa = 4.43KK13 pKa = 10.62LSGASFLAVATRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84QDD30 pKa = 3.05LSFLEE35 pKa = 4.52GQLDD39 pKa = 3.38KK40 pKa = 10.59CTRR43 pKa = 11.84LSPQFHH49 pKa = 6.55LPIRR53 pKa = 11.84DD54 pKa = 3.24IPEE57 pKa = 3.87IKK59 pKa = 9.44STGKK63 pKa = 11.03GDD65 pKa = 3.41MQRR68 pKa = 11.84AKK70 pKa = 9.41TQTFDD75 pKa = 3.82VPDD78 pKa = 4.05NFEE81 pKa = 4.59KK82 pKa = 10.88LPPIPNSKK90 pKa = 8.61LTRR93 pKa = 11.84NGEE96 pKa = 4.3LIQKK100 pKa = 9.97GNLGKK105 pKa = 10.37RR106 pKa = 11.84FYY108 pKa = 11.09NSQKK112 pKa = 9.23TFGGKK117 pKa = 8.8NVPDD121 pKa = 4.13NLRR124 pKa = 11.84QTIRR128 pKa = 11.84HH129 pKa = 5.59ALATRR134 pKa = 11.84IEE136 pKa = 4.38INGPAVEE143 pKa = 4.39SCVIADD149 pKa = 3.71EE150 pKa = 4.78LMATGLEE157 pKa = 4.2WEE159 pKa = 4.59YY160 pKa = 11.59SAGTTQGLVEE170 pKa = 4.02RR171 pKa = 11.84LSRR174 pKa = 11.84FGKK177 pKa = 10.05DD178 pKa = 2.92DD179 pKa = 3.29KK180 pKa = 10.83TVSFFRR186 pKa = 11.84QTDD189 pKa = 3.91EE190 pKa = 3.57IRR192 pKa = 11.84NFQSFLFQALDD203 pKa = 3.28TLLPLKK209 pKa = 9.03STPFEE214 pKa = 3.91TSFPSTSTEE223 pKa = 3.9VIEE226 pKa = 4.97GIRR229 pKa = 11.84VNPQSSAGPMYY240 pKa = 9.96GTTKK244 pKa = 10.39VQALDD249 pKa = 5.17DD250 pKa = 3.73IMLTMDD256 pKa = 5.59AIRR259 pKa = 11.84TQFSLGTLDD268 pKa = 3.53TFFAAHH274 pKa = 7.51PEE276 pKa = 4.04TLIADD281 pKa = 4.28CRR283 pKa = 11.84NKK285 pKa = 9.38TDD287 pKa = 3.18RR288 pKa = 11.84YY289 pKa = 9.91EE290 pKa = 3.98IAKK293 pKa = 10.14LPEE296 pKa = 3.66KK297 pKa = 9.66TRR299 pKa = 11.84PYY301 pKa = 10.33WSFNAHH307 pKa = 3.89WQFYY311 pKa = 8.49YY312 pKa = 10.41SWLMQRR318 pKa = 11.84FCDD321 pKa = 3.84NLYY324 pKa = 9.98VASDD328 pKa = 4.16PEE330 pKa = 4.95CDD332 pKa = 3.31VPTYY336 pKa = 10.94NYY338 pKa = 9.99YY339 pKa = 10.07GHH341 pKa = 6.53VWAKK345 pKa = 10.64GGAQRR350 pKa = 11.84IHH352 pKa = 6.17EE353 pKa = 4.83SIMNVAKK360 pKa = 10.01TGRR363 pKa = 11.84PFVGIYY369 pKa = 10.82GDD371 pKa = 4.08DD372 pKa = 3.63FLFAFPIKK380 pKa = 9.4TDD382 pKa = 3.25EE383 pKa = 4.27GTTIQVMAPDD393 pKa = 4.43ISQMDD398 pKa = 3.94STVSHH403 pKa = 5.93AVIRR407 pKa = 11.84GTCDD411 pKa = 3.23YY412 pKa = 10.54IYY414 pKa = 10.71KK415 pKa = 10.12QYY417 pKa = 11.08QEE419 pKa = 4.29AHH421 pKa = 6.44GDD423 pKa = 3.65DD424 pKa = 3.76PFVRR428 pKa = 11.84FLLTRR433 pKa = 11.84LAEE436 pKa = 4.57DD437 pKa = 3.34ASSPRR442 pKa = 11.84FLASAIATYY451 pKa = 10.49QNQRR455 pKa = 11.84PTGLATGVVGTTLFDD470 pKa = 3.64TVQAALLAFSVIEE483 pKa = 3.68EE484 pKa = 4.11WNRR487 pKa = 11.84TKK489 pKa = 10.84RR490 pKa = 11.84SPDD493 pKa = 3.23EE494 pKa = 3.99IFNKK498 pKa = 9.52VSKK501 pKa = 10.43KK502 pKa = 10.02FGMRR506 pKa = 11.84AKK508 pKa = 10.42SLEE511 pKa = 3.94LYY513 pKa = 9.45EE514 pKa = 4.55YY515 pKa = 10.94NGLPPAEE522 pKa = 4.08GTEE525 pKa = 4.08WTFLGMQLATYY536 pKa = 10.49SNSKK540 pKa = 7.91HH541 pKa = 5.35TNGVIPRR548 pKa = 11.84LPRR551 pKa = 11.84DD552 pKa = 3.56SLLKK556 pKa = 10.72SILNPRR562 pKa = 11.84INSYY566 pKa = 10.77LFSSKK571 pKa = 10.64LGTQRR576 pKa = 11.84YY577 pKa = 8.91LFDD580 pKa = 5.0ASRR583 pKa = 11.84GWYY586 pKa = 9.86LCALGDD592 pKa = 3.78RR593 pKa = 11.84LIWDD597 pKa = 3.73VLSSVIEE604 pKa = 4.27TVPTMAVGMVVQAGGGLGAPPEE626 pKa = 4.46CNGLFDD632 pKa = 4.79VNRR635 pKa = 11.84PFNWLSSFGVPTMDD649 pKa = 2.77WVADD653 pKa = 4.05LFLSPMDD660 pKa = 4.97PDD662 pKa = 3.78PAGWLSLLDD671 pKa = 3.86PPKK674 pKa = 10.78GRR676 pKa = 11.84LIVEE680 pKa = 4.29MQDD683 pKa = 3.26QVDD686 pKa = 4.11LSRR689 pKa = 11.84YY690 pKa = 6.89TIRR693 pKa = 11.84PTPNTAPHH701 pKa = 5.44SWEE704 pKa = 3.81TRR706 pKa = 11.84EE707 pKa = 4.99KK708 pKa = 10.13VTDD711 pKa = 3.62TNLEE715 pKa = 4.19SLVSDD720 pKa = 3.88NPSGLGPQVQRR731 pKa = 11.84PKK733 pKa = 10.51AQHH736 pKa = 6.23LPPVKK741 pKa = 10.41APPFKK746 pKa = 10.41IRR748 pKa = 11.84LSEE751 pKa = 4.1HH752 pKa = 6.27LSKK755 pKa = 11.18HH756 pKa = 5.96NITAFVPSEE765 pKa = 3.76LAAEE769 pKa = 4.53LGVSVTTILTEE780 pKa = 4.41LYY782 pKa = 10.45SKK784 pKa = 10.52EE785 pKa = 4.33RR786 pKa = 11.84EE787 pKa = 4.06TKK789 pKa = 9.72APEE792 pKa = 3.78FRR794 pKa = 11.84WYY796 pKa = 10.51SLGRR800 pKa = 11.84ILTKK804 pKa = 10.9DD805 pKa = 3.29PVDD808 pKa = 3.55TVQPSRR814 pKa = 11.84ILWNQYY820 pKa = 9.05SRR822 pKa = 11.84ILSEE826 pKa = 4.63LGYY829 pKa = 10.34SNPAIQPARR838 pKa = 11.84PKK840 pKa = 10.27AGPVPAPRR848 pKa = 11.84YY849 pKa = 10.25SKK851 pKa = 10.26MKK853 pKa = 10.78ANVPQDD859 pKa = 3.47FATSNSTTISDD870 pKa = 4.63CDD872 pKa = 3.4TSPAIRR878 pKa = 11.84VSGPDD883 pKa = 3.23QTPTSQTVPQYY894 pKa = 9.79TPKK897 pKa = 9.26PAQMMYY903 pKa = 10.64QRR905 pKa = 11.84IPAKK909 pKa = 9.83TGAVVCALEE918 pKa = 4.36EE919 pKa = 4.08NLVAPRR925 pKa = 11.84AEE927 pKa = 4.3GLSSGARR934 pKa = 11.84VPLADD939 pKa = 3.46QVNSGRR945 pKa = 11.84IVHH948 pKa = 6.36QYY950 pKa = 10.35FNWDD954 pKa = 3.57PPSPILSTSPEE965 pKa = 3.76QTLPLDD971 pKa = 4.23LSTSEE976 pKa = 4.65SVSTPDD982 pKa = 3.19TTSTPDD988 pKa = 2.96TVMYY992 pKa = 9.92RR993 pKa = 11.84SPPIEE998 pKa = 4.25GSQCYY1003 pKa = 9.95EE1004 pKa = 3.96EE1005 pKa = 4.8DD1006 pKa = 3.38QSDD1009 pKa = 3.84GSEE1012 pKa = 4.29SSWTSYY1018 pKa = 10.88ASSVSGCQRR1027 pKa = 11.84FDD1029 pKa = 3.37YY1030 pKa = 10.32PNRR1033 pKa = 11.84RR1034 pKa = 11.84SHH1036 pKa = 7.51PDD1038 pKa = 2.9AFPNQEE1044 pKa = 3.94AQSFRR1049 pKa = 11.84AAKK1052 pKa = 8.96EE1053 pKa = 3.46QYY1055 pKa = 9.72EE1056 pKa = 4.56RR1057 pKa = 11.84YY1058 pKa = 7.56TEE1060 pKa = 4.01SRR1062 pKa = 11.84RR1063 pKa = 11.84TQSPPQSPPPRR1074 pKa = 11.84WPTPPPPIPITEE1086 pKa = 4.3GSLTYY1091 pKa = 10.23TSVSTTTSNSKK1102 pKa = 9.97QVASGDD1108 pKa = 3.47RR1109 pKa = 11.84SRR1111 pKa = 11.84GSGAKK1116 pKa = 9.59PKK1118 pKa = 10.68VSTHH1122 pKa = 5.33SKK1124 pKa = 10.43KK1125 pKa = 9.9STDD1128 pKa = 3.36VPSQVPVKK1136 pKa = 10.2SVEE1139 pKa = 4.1VKK1141 pKa = 10.59GRR1143 pKa = 11.84TFTNSTRR1150 pKa = 11.84FSGVEE1155 pKa = 3.79HH1156 pKa = 6.8KK1157 pKa = 10.07PVKK1160 pKa = 7.75TTNGNQRR1167 pKa = 11.84VFYY1170 pKa = 10.0KK1171 pKa = 10.82VKK1173 pKa = 10.69SKK1175 pKa = 10.94DD1176 pKa = 3.37FRR1178 pKa = 11.84HH1179 pKa = 6.24PPVPPTIVGFYY1190 pKa = 10.21DD1191 pKa = 3.98GKK1193 pKa = 10.22HH1194 pKa = 5.39NYY1196 pKa = 8.57YY1197 pKa = 10.44VPTYY1201 pKa = 9.69KK1202 pKa = 10.61CQVDD1206 pKa = 3.95SKK1208 pKa = 9.22VKK1210 pKa = 10.5CVNTPPNPTKK1220 pKa = 10.62LGNFAWLGSGVSSLCAWAVTGKK1242 pKa = 10.64FEE1244 pKa = 5.13LVFDD1248 pKa = 4.95FTHH1251 pKa = 6.51QPDD1254 pKa = 3.66KK1255 pKa = 11.05MLFQCSIYY1263 pKa = 10.06IDD1265 pKa = 3.87GKK1267 pKa = 11.32LMISASRR1274 pKa = 11.84CKK1276 pKa = 10.2SKK1278 pKa = 11.16KK1279 pKa = 7.04EE1280 pKa = 3.95AKK1282 pKa = 9.91EE1283 pKa = 3.3IAAFQFLHH1291 pKa = 6.15SCFLHH1296 pKa = 5.22RR1297 pKa = 11.84TPRR1300 pKa = 11.84DD1301 pKa = 3.88LIHH1304 pKa = 6.85HH1305 pKa = 6.67ALKK1308 pKa = 9.84TPHH1311 pKa = 6.49
Molecular weight: 146.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1978 |
667 |
1311 |
989.0 |
109.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.269 ± 0.212 | 1.163 ± 0.207 |
5.561 ± 0.219 | 4.601 ± 0.202 |
4.348 ± 0.301 | 5.966 ± 0.242 |
1.517 ± 0.385 | 4.752 ± 0.55 |
4.954 ± 0.38 | 7.179 ± 0.368 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.517 ± 0.235 | 3.488 ± 0.246 |
7.836 ± 0.547 | 4.398 ± 0.176 |
5.308 ± 0.407 | 10.111 ± 0.719 |
9.808 ± 1.7 | 6.32 ± 0.214 |
1.314 ± 0.208 | 3.589 ± 0.23 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
