
Torque teno virus 23
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 7.38
Get precalculated fractions of proteins
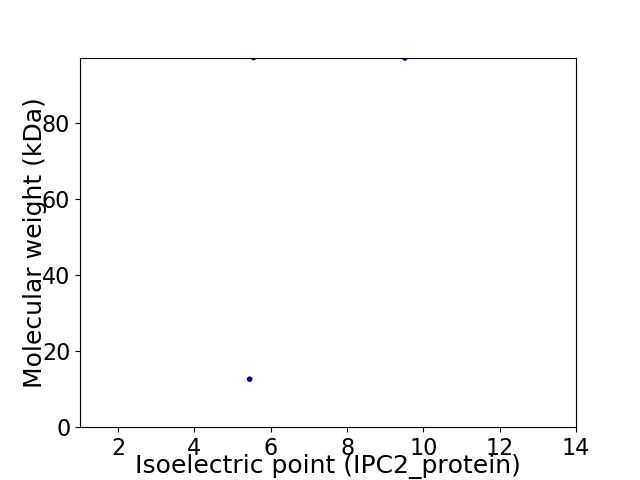
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9DTD5|Q9DTD5_9VIRU ORF2 ORF1 genes isolate: s-TTV CH65-2 OS=Torque teno virus 23 OX=687362 PE=4 SV=1
MM1 pKa = 7.45SLTPPAHH8 pKa = 5.79NASGIEE14 pKa = 4.11RR15 pKa = 11.84NWFEE19 pKa = 4.29SCFRR23 pKa = 11.84SHH25 pKa = 7.21AAVCGCGHH33 pKa = 6.79FVNHH37 pKa = 6.56LNVPAARR44 pKa = 11.84YY45 pKa = 8.94AFNGGPTPPGGPRR58 pKa = 11.84SGPPQIRR65 pKa = 11.84RR66 pKa = 11.84ALPAPAPPEE75 pKa = 4.29PPAHH79 pKa = 5.77NPWRR83 pKa = 11.84GGDD86 pKa = 3.39AGDD89 pKa = 3.36AAGRR93 pKa = 11.84GGGRR97 pKa = 11.84DD98 pKa = 3.2AGAGGGEE105 pKa = 4.17YY106 pKa = 10.6APEE109 pKa = 3.93EE110 pKa = 4.15LDD112 pKa = 3.58ALFAAVEE119 pKa = 4.25EE120 pKa = 4.6DD121 pKa = 3.79TQQ123 pKa = 3.8
MM1 pKa = 7.45SLTPPAHH8 pKa = 5.79NASGIEE14 pKa = 4.11RR15 pKa = 11.84NWFEE19 pKa = 4.29SCFRR23 pKa = 11.84SHH25 pKa = 7.21AAVCGCGHH33 pKa = 6.79FVNHH37 pKa = 6.56LNVPAARR44 pKa = 11.84YY45 pKa = 8.94AFNGGPTPPGGPRR58 pKa = 11.84SGPPQIRR65 pKa = 11.84RR66 pKa = 11.84ALPAPAPPEE75 pKa = 4.29PPAHH79 pKa = 5.77NPWRR83 pKa = 11.84GGDD86 pKa = 3.39AGDD89 pKa = 3.36AAGRR93 pKa = 11.84GGGRR97 pKa = 11.84DD98 pKa = 3.2AGAGGGEE105 pKa = 4.17YY106 pKa = 10.6APEE109 pKa = 3.93EE110 pKa = 4.15LDD112 pKa = 3.58ALFAAVEE119 pKa = 4.25EE120 pKa = 4.6DD121 pKa = 3.79TQQ123 pKa = 3.8
Molecular weight: 12.62 kDa
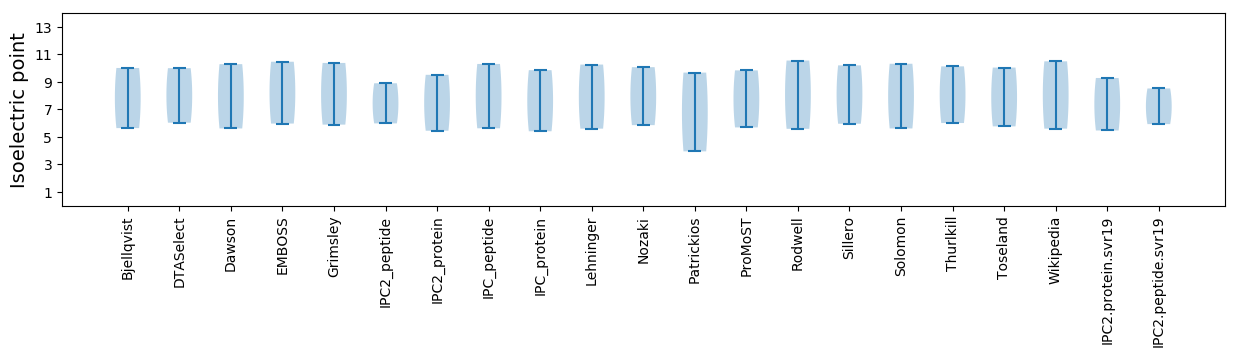
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9DTD5|Q9DTD5_9VIRU ORF2 ORF1 genes isolate: s-TTV CH65-2 OS=Torque teno virus 23 OX=687362 PE=4 SV=1
MM1 pKa = 7.27ARR3 pKa = 11.84WRR5 pKa = 11.84RR6 pKa = 11.84WRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84PWWWKK16 pKa = 7.69RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84WRR23 pKa = 11.84RR24 pKa = 11.84VRR26 pKa = 11.84PRR28 pKa = 11.84RR29 pKa = 11.84ARR31 pKa = 11.84RR32 pKa = 11.84TVRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84YY41 pKa = 5.75TVRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84WGRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84GRR54 pKa = 11.84LRR56 pKa = 11.84RR57 pKa = 11.84TRR59 pKa = 11.84WRR61 pKa = 11.84RR62 pKa = 11.84TRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 9.66RR69 pKa = 11.84KK70 pKa = 7.66TLILRR75 pKa = 11.84QWNPATVRR83 pKa = 11.84SCRR86 pKa = 11.84IKK88 pKa = 10.86GLVPMVICGHH98 pKa = 5.23TRR100 pKa = 11.84AGRR103 pKa = 11.84NYY105 pKa = 10.56AMHH108 pKa = 6.65SEE110 pKa = 4.71DD111 pKa = 3.64YY112 pKa = 9.94PEE114 pKa = 3.84QGRR117 pKa = 11.84YY118 pKa = 8.76PFGGSLSTTTWSLKK132 pKa = 10.21VLFDD136 pKa = 3.39EE137 pKa = 4.86HH138 pKa = 7.37QKK140 pKa = 10.25FHH142 pKa = 6.2NRR144 pKa = 11.84WSRR147 pKa = 11.84SNQDD151 pKa = 2.8LDD153 pKa = 3.51LARR156 pKa = 11.84YY157 pKa = 8.46LGCRR161 pKa = 11.84FTFYY165 pKa = 10.5RR166 pKa = 11.84DD167 pKa = 2.85KK168 pKa = 10.43KK169 pKa = 7.65TDD171 pKa = 3.67YY172 pKa = 10.84IVTYY176 pKa = 10.36RR177 pKa = 11.84NKK179 pKa = 10.49PPFKK183 pKa = 10.09INKK186 pKa = 8.58LDD188 pKa = 3.52SPMSHH193 pKa = 7.52PGMLMQQKK201 pKa = 9.61QKK203 pKa = 10.78IIIPSYY209 pKa = 8.12DD210 pKa = 3.32TRR212 pKa = 11.84PGGPQKK218 pKa = 9.54ITVKK222 pKa = 10.07IRR224 pKa = 11.84PPKK227 pKa = 10.07LFQDD231 pKa = 3.01KK232 pKa = 9.89WYY234 pKa = 8.34TQTDD238 pKa = 3.69LCEE241 pKa = 4.12VNLVSFVVSACSLTHH256 pKa = 6.41PFCPPLTNNPCVTFQVLKK274 pKa = 10.77DD275 pKa = 4.05FYY277 pKa = 11.27YY278 pKa = 11.25GNLNIAEE285 pKa = 4.4KK286 pKa = 9.9TNSKK290 pKa = 11.0NIFEE294 pKa = 4.59TLKK297 pKa = 10.49KK298 pKa = 10.22HH299 pKa = 5.68PGYY302 pKa = 10.64YY303 pKa = 10.34EE304 pKa = 4.07NFLTQTLLLGDD315 pKa = 4.22VKK317 pKa = 10.33PAKK320 pKa = 10.9YY321 pKa = 10.53NDD323 pKa = 3.86TNGSLKK329 pKa = 9.9STITNASIKK338 pKa = 9.51TNNNGQFGMYY348 pKa = 10.1VYY350 pKa = 11.01DD351 pKa = 3.94KK352 pKa = 10.84DD353 pKa = 3.84KK354 pKa = 11.5LGAALQKK361 pKa = 10.6AVEE364 pKa = 4.68WYY366 pKa = 7.88HH367 pKa = 5.8QQYY370 pKa = 7.58QTHH373 pKa = 6.59NDD375 pKa = 2.83VHH377 pKa = 7.9SIYY380 pKa = 10.79GKK382 pKa = 8.4PTNEE386 pKa = 3.49YY387 pKa = 10.02LGYY390 pKa = 8.68HH391 pKa = 5.58TGIYY395 pKa = 10.24SPMVLSPMRR404 pKa = 11.84SNLDD408 pKa = 3.25MEE410 pKa = 5.33RR411 pKa = 11.84AFQDD415 pKa = 3.07VTYY418 pKa = 10.91NPNADD423 pKa = 3.52RR424 pKa = 11.84GKK426 pKa = 11.38GNIVWFQYY434 pKa = 7.35STRR437 pKa = 11.84NDD439 pKa = 3.08TTFDD443 pKa = 3.27EE444 pKa = 4.5KK445 pKa = 10.94QYY447 pKa = 11.4KK448 pKa = 9.81CVLQDD453 pKa = 3.51LPIWAALYY461 pKa = 10.01GYY463 pKa = 10.58SDD465 pKa = 3.72FVEE468 pKa = 5.17AEE470 pKa = 4.1LGISAEE476 pKa = 3.96IHH478 pKa = 5.6NSGVIVVVCPTLSTNEE494 pKa = 4.07KK495 pKa = 8.73GTIGAQIQGYY505 pKa = 9.42IFYY508 pKa = 8.04DD509 pKa = 3.45TLFGQGEE516 pKa = 4.25MPDD519 pKa = 3.84GRR521 pKa = 11.84GHH523 pKa = 7.16IPIYY527 pKa = 8.86WQSRR531 pKa = 11.84WYY533 pKa = 9.46PRR535 pKa = 11.84SSFQQQVIHH544 pKa = 7.15DD545 pKa = 4.35PVLSGPFSYY554 pKa = 10.98KK555 pKa = 10.39DD556 pKa = 3.59DD557 pKa = 4.25LVSTTLTAAYY567 pKa = 9.99SFKK570 pKa = 10.63FLWGGDD576 pKa = 3.87TISEE580 pKa = 3.99QVVRR584 pKa = 11.84NPCRR588 pKa = 11.84ADD590 pKa = 3.4APAPSYY596 pKa = 9.22PHH598 pKa = 6.4RR599 pKa = 11.84QPRR602 pKa = 11.84DD603 pKa = 3.47VQVIDD608 pKa = 3.87PRR610 pKa = 11.84LVGPQWVFHH619 pKa = 5.21TWDD622 pKa = 2.84WRR624 pKa = 11.84RR625 pKa = 11.84GLFGKK630 pKa = 10.31KK631 pKa = 9.57AIKK634 pKa = 10.0RR635 pKa = 11.84VSQKK639 pKa = 10.57PDD641 pKa = 3.38DD642 pKa = 5.36DD643 pKa = 4.73EE644 pKa = 7.08DD645 pKa = 3.59FTAPYY650 pKa = 9.67KK651 pKa = 10.11IPRR654 pKa = 11.84YY655 pKa = 9.11FPPPDD660 pKa = 3.2RR661 pKa = 11.84EE662 pKa = 4.21GARR665 pKa = 11.84EE666 pKa = 3.83EE667 pKa = 4.78DD668 pKa = 3.73SASEE672 pKa = 4.82SEE674 pKa = 4.25DD675 pKa = 3.11QGSNFIPEE683 pKa = 3.99EE684 pKa = 4.12EE685 pKa = 4.19EE686 pKa = 3.72ALEE689 pKa = 4.76AGLQGSQLQQHH700 pKa = 5.91RR701 pKa = 11.84VQRR704 pKa = 11.84VYY706 pKa = 10.72ILQQLRR712 pKa = 11.84EE713 pKa = 4.14QRR715 pKa = 11.84RR716 pKa = 11.84MGQQLQHH723 pKa = 6.3LTRR726 pKa = 11.84EE727 pKa = 4.01LLKK730 pKa = 10.5RR731 pKa = 11.84RR732 pKa = 11.84RR733 pKa = 11.84LQYY736 pKa = 10.35KK737 pKa = 8.16PRR739 pKa = 11.84NYY741 pKa = 9.84FPKK744 pKa = 9.98LRR746 pKa = 11.84QRR748 pKa = 11.84YY749 pKa = 6.19TCFLHH754 pKa = 7.32LDD756 pKa = 3.88PDD758 pKa = 3.97ADD760 pKa = 3.7ALPRR764 pKa = 11.84KK765 pKa = 9.44SGKK768 pKa = 9.66PNTRR772 pKa = 11.84PAEE775 pKa = 4.21PSTAPLEE782 pKa = 4.44PTSTAPLCYY791 pKa = 9.26PYY793 pKa = 10.55LPSDD797 pKa = 3.64VCVKK801 pKa = 9.83FEE803 pKa = 4.41PQLQINKK810 pKa = 9.71AVGVTTWSVSVYY822 pKa = 10.24KK823 pKa = 10.57SHH825 pKa = 7.38
MM1 pKa = 7.27ARR3 pKa = 11.84WRR5 pKa = 11.84RR6 pKa = 11.84WRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84PWWWKK16 pKa = 7.69RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84WRR23 pKa = 11.84RR24 pKa = 11.84VRR26 pKa = 11.84PRR28 pKa = 11.84RR29 pKa = 11.84ARR31 pKa = 11.84RR32 pKa = 11.84TVRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84YY41 pKa = 5.75TVRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84WGRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84GRR54 pKa = 11.84LRR56 pKa = 11.84RR57 pKa = 11.84TRR59 pKa = 11.84WRR61 pKa = 11.84RR62 pKa = 11.84TRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 9.66RR69 pKa = 11.84KK70 pKa = 7.66TLILRR75 pKa = 11.84QWNPATVRR83 pKa = 11.84SCRR86 pKa = 11.84IKK88 pKa = 10.86GLVPMVICGHH98 pKa = 5.23TRR100 pKa = 11.84AGRR103 pKa = 11.84NYY105 pKa = 10.56AMHH108 pKa = 6.65SEE110 pKa = 4.71DD111 pKa = 3.64YY112 pKa = 9.94PEE114 pKa = 3.84QGRR117 pKa = 11.84YY118 pKa = 8.76PFGGSLSTTTWSLKK132 pKa = 10.21VLFDD136 pKa = 3.39EE137 pKa = 4.86HH138 pKa = 7.37QKK140 pKa = 10.25FHH142 pKa = 6.2NRR144 pKa = 11.84WSRR147 pKa = 11.84SNQDD151 pKa = 2.8LDD153 pKa = 3.51LARR156 pKa = 11.84YY157 pKa = 8.46LGCRR161 pKa = 11.84FTFYY165 pKa = 10.5RR166 pKa = 11.84DD167 pKa = 2.85KK168 pKa = 10.43KK169 pKa = 7.65TDD171 pKa = 3.67YY172 pKa = 10.84IVTYY176 pKa = 10.36RR177 pKa = 11.84NKK179 pKa = 10.49PPFKK183 pKa = 10.09INKK186 pKa = 8.58LDD188 pKa = 3.52SPMSHH193 pKa = 7.52PGMLMQQKK201 pKa = 9.61QKK203 pKa = 10.78IIIPSYY209 pKa = 8.12DD210 pKa = 3.32TRR212 pKa = 11.84PGGPQKK218 pKa = 9.54ITVKK222 pKa = 10.07IRR224 pKa = 11.84PPKK227 pKa = 10.07LFQDD231 pKa = 3.01KK232 pKa = 9.89WYY234 pKa = 8.34TQTDD238 pKa = 3.69LCEE241 pKa = 4.12VNLVSFVVSACSLTHH256 pKa = 6.41PFCPPLTNNPCVTFQVLKK274 pKa = 10.77DD275 pKa = 4.05FYY277 pKa = 11.27YY278 pKa = 11.25GNLNIAEE285 pKa = 4.4KK286 pKa = 9.9TNSKK290 pKa = 11.0NIFEE294 pKa = 4.59TLKK297 pKa = 10.49KK298 pKa = 10.22HH299 pKa = 5.68PGYY302 pKa = 10.64YY303 pKa = 10.34EE304 pKa = 4.07NFLTQTLLLGDD315 pKa = 4.22VKK317 pKa = 10.33PAKK320 pKa = 10.9YY321 pKa = 10.53NDD323 pKa = 3.86TNGSLKK329 pKa = 9.9STITNASIKK338 pKa = 9.51TNNNGQFGMYY348 pKa = 10.1VYY350 pKa = 11.01DD351 pKa = 3.94KK352 pKa = 10.84DD353 pKa = 3.84KK354 pKa = 11.5LGAALQKK361 pKa = 10.6AVEE364 pKa = 4.68WYY366 pKa = 7.88HH367 pKa = 5.8QQYY370 pKa = 7.58QTHH373 pKa = 6.59NDD375 pKa = 2.83VHH377 pKa = 7.9SIYY380 pKa = 10.79GKK382 pKa = 8.4PTNEE386 pKa = 3.49YY387 pKa = 10.02LGYY390 pKa = 8.68HH391 pKa = 5.58TGIYY395 pKa = 10.24SPMVLSPMRR404 pKa = 11.84SNLDD408 pKa = 3.25MEE410 pKa = 5.33RR411 pKa = 11.84AFQDD415 pKa = 3.07VTYY418 pKa = 10.91NPNADD423 pKa = 3.52RR424 pKa = 11.84GKK426 pKa = 11.38GNIVWFQYY434 pKa = 7.35STRR437 pKa = 11.84NDD439 pKa = 3.08TTFDD443 pKa = 3.27EE444 pKa = 4.5KK445 pKa = 10.94QYY447 pKa = 11.4KK448 pKa = 9.81CVLQDD453 pKa = 3.51LPIWAALYY461 pKa = 10.01GYY463 pKa = 10.58SDD465 pKa = 3.72FVEE468 pKa = 5.17AEE470 pKa = 4.1LGISAEE476 pKa = 3.96IHH478 pKa = 5.6NSGVIVVVCPTLSTNEE494 pKa = 4.07KK495 pKa = 8.73GTIGAQIQGYY505 pKa = 9.42IFYY508 pKa = 8.04DD509 pKa = 3.45TLFGQGEE516 pKa = 4.25MPDD519 pKa = 3.84GRR521 pKa = 11.84GHH523 pKa = 7.16IPIYY527 pKa = 8.86WQSRR531 pKa = 11.84WYY533 pKa = 9.46PRR535 pKa = 11.84SSFQQQVIHH544 pKa = 7.15DD545 pKa = 4.35PVLSGPFSYY554 pKa = 10.98KK555 pKa = 10.39DD556 pKa = 3.59DD557 pKa = 4.25LVSTTLTAAYY567 pKa = 9.99SFKK570 pKa = 10.63FLWGGDD576 pKa = 3.87TISEE580 pKa = 3.99QVVRR584 pKa = 11.84NPCRR588 pKa = 11.84ADD590 pKa = 3.4APAPSYY596 pKa = 9.22PHH598 pKa = 6.4RR599 pKa = 11.84QPRR602 pKa = 11.84DD603 pKa = 3.47VQVIDD608 pKa = 3.87PRR610 pKa = 11.84LVGPQWVFHH619 pKa = 5.21TWDD622 pKa = 2.84WRR624 pKa = 11.84RR625 pKa = 11.84GLFGKK630 pKa = 10.31KK631 pKa = 9.57AIKK634 pKa = 10.0RR635 pKa = 11.84VSQKK639 pKa = 10.57PDD641 pKa = 3.38DD642 pKa = 5.36DD643 pKa = 4.73EE644 pKa = 7.08DD645 pKa = 3.59FTAPYY650 pKa = 9.67KK651 pKa = 10.11IPRR654 pKa = 11.84YY655 pKa = 9.11FPPPDD660 pKa = 3.2RR661 pKa = 11.84EE662 pKa = 4.21GARR665 pKa = 11.84EE666 pKa = 3.83EE667 pKa = 4.78DD668 pKa = 3.73SASEE672 pKa = 4.82SEE674 pKa = 4.25DD675 pKa = 3.11QGSNFIPEE683 pKa = 3.99EE684 pKa = 4.12EE685 pKa = 4.19EE686 pKa = 3.72ALEE689 pKa = 4.76AGLQGSQLQQHH700 pKa = 5.91RR701 pKa = 11.84VQRR704 pKa = 11.84VYY706 pKa = 10.72ILQQLRR712 pKa = 11.84EE713 pKa = 4.14QRR715 pKa = 11.84RR716 pKa = 11.84MGQQLQHH723 pKa = 6.3LTRR726 pKa = 11.84EE727 pKa = 4.01LLKK730 pKa = 10.5RR731 pKa = 11.84RR732 pKa = 11.84RR733 pKa = 11.84LQYY736 pKa = 10.35KK737 pKa = 8.16PRR739 pKa = 11.84NYY741 pKa = 9.84FPKK744 pKa = 9.98LRR746 pKa = 11.84QRR748 pKa = 11.84YY749 pKa = 6.19TCFLHH754 pKa = 7.32LDD756 pKa = 3.88PDD758 pKa = 3.97ADD760 pKa = 3.7ALPRR764 pKa = 11.84KK765 pKa = 9.44SGKK768 pKa = 9.66PNTRR772 pKa = 11.84PAEE775 pKa = 4.21PSTAPLEE782 pKa = 4.44PTSTAPLCYY791 pKa = 9.26PYY793 pKa = 10.55LPSDD797 pKa = 3.64VCVKK801 pKa = 9.83FEE803 pKa = 4.41PQLQINKK810 pKa = 9.71AVGVTTWSVSVYY822 pKa = 10.24KK823 pKa = 10.57SHH825 pKa = 7.38
Molecular weight: 97.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
948 |
123 |
825 |
474.0 |
54.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.013 ± 4.781 | 1.688 ± 0.351 |
5.169 ± 0.515 | 4.325 ± 1.017 |
3.903 ± 0.076 | 6.962 ± 3.959 |
2.637 ± 0.666 | 3.797 ± 1.013 |
5.169 ± 2.412 | 6.857 ± 1.303 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.371 ± 0.261 | 4.219 ± 0.307 |
8.017 ± 2.708 | 5.38 ± 1.751 |
10.338 ± 1.409 | 5.591 ± 0.712 |
5.907 ± 1.618 | 5.274 ± 0.944 |
2.532 ± 0.423 | 4.852 ± 1.505 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
