
Salibacterium qingdaonense
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Salibacterium
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
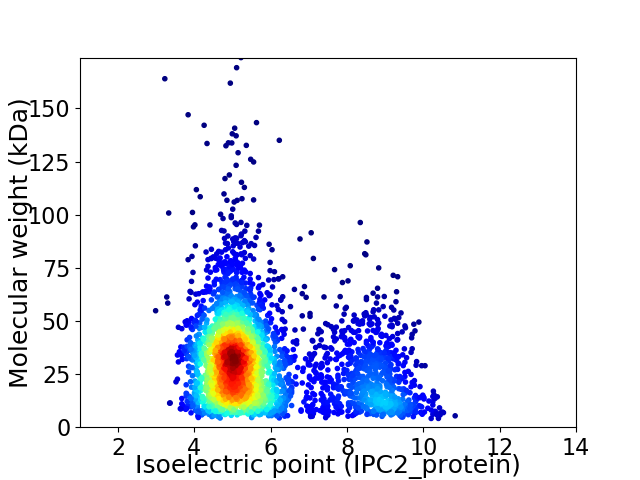
Virtual 2D-PAGE plot for 3532 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I4PXI8|A0A1I4PXI8_9BACI Two-component system CitB family response regulator MalR OS=Salibacterium qingdaonense OX=266892 GN=SAMN04488054_13316 PE=4 SV=1
MM1 pKa = 7.18FKK3 pKa = 10.24KK4 pKa = 10.51VSVFIIVSFALLLSACGSSGSSSEE28 pKa = 4.4EE29 pKa = 3.67NASNYY34 pKa = 9.71PEE36 pKa = 3.99NQIEE40 pKa = 4.55VIVPFSSGGASDD52 pKa = 4.05LVSRR56 pKa = 11.84TVASEE61 pKa = 3.94MEE63 pKa = 4.17NDD65 pKa = 3.6LDD67 pKa = 4.5VPLTITNQTGGSGATGMLSLKK88 pKa = 10.04NAQADD93 pKa = 4.78GYY95 pKa = 8.4TIGYY99 pKa = 8.9VPVEE103 pKa = 3.92MSMLDD108 pKa = 3.62SLEE111 pKa = 4.4LADD114 pKa = 4.54ITPSDD119 pKa = 4.06YY120 pKa = 11.49EE121 pKa = 4.51FIGQLMTIPSAITVPADD138 pKa = 3.2APYY141 pKa = 9.23DD142 pKa = 3.81TIEE145 pKa = 4.26EE146 pKa = 4.22FVSYY150 pKa = 11.09AEE152 pKa = 4.29EE153 pKa = 4.28NPGEE157 pKa = 4.05IQIGNSGTGSIWHH170 pKa = 6.63IAAAAFAEE178 pKa = 4.34EE179 pKa = 4.06AGIDD183 pKa = 3.95VEE185 pKa = 4.34YY186 pKa = 11.02VPYY189 pKa = 10.69DD190 pKa = 3.56GASPAVTALMGGHH203 pKa = 6.81ISAVSVSPSEE213 pKa = 3.87VRR215 pKa = 11.84GGVEE219 pKa = 4.11SGDD222 pKa = 3.73LKK224 pKa = 11.23VLGVMGEE231 pKa = 4.07EE232 pKa = 4.58RR233 pKa = 11.84DD234 pKa = 3.84PEE236 pKa = 4.7LPDD239 pKa = 3.3VPTMQEE245 pKa = 2.98AGYY248 pKa = 9.72DD249 pKa = 3.54VSVAGWGGFVAPEE262 pKa = 4.02GTPEE266 pKa = 3.99EE267 pKa = 4.16VLEE270 pKa = 4.16PLRR273 pKa = 11.84SSFKK277 pKa = 10.38NAAEE281 pKa = 4.08SEE283 pKa = 4.32EE284 pKa = 4.22FQQLMDD290 pKa = 4.45DD291 pKa = 3.75RR292 pKa = 11.84GMIPAYY298 pKa = 9.96KK299 pKa = 9.89NGKK302 pKa = 8.57EE303 pKa = 3.85FSSYY307 pKa = 11.39AEE309 pKa = 3.67EE310 pKa = 4.22QYY312 pKa = 11.43DD313 pKa = 4.04YY314 pKa = 10.87FSEE317 pKa = 5.1LIPTIEE323 pKa = 4.13INQQ326 pKa = 3.46
MM1 pKa = 7.18FKK3 pKa = 10.24KK4 pKa = 10.51VSVFIIVSFALLLSACGSSGSSSEE28 pKa = 4.4EE29 pKa = 3.67NASNYY34 pKa = 9.71PEE36 pKa = 3.99NQIEE40 pKa = 4.55VIVPFSSGGASDD52 pKa = 4.05LVSRR56 pKa = 11.84TVASEE61 pKa = 3.94MEE63 pKa = 4.17NDD65 pKa = 3.6LDD67 pKa = 4.5VPLTITNQTGGSGATGMLSLKK88 pKa = 10.04NAQADD93 pKa = 4.78GYY95 pKa = 8.4TIGYY99 pKa = 8.9VPVEE103 pKa = 3.92MSMLDD108 pKa = 3.62SLEE111 pKa = 4.4LADD114 pKa = 4.54ITPSDD119 pKa = 4.06YY120 pKa = 11.49EE121 pKa = 4.51FIGQLMTIPSAITVPADD138 pKa = 3.2APYY141 pKa = 9.23DD142 pKa = 3.81TIEE145 pKa = 4.26EE146 pKa = 4.22FVSYY150 pKa = 11.09AEE152 pKa = 4.29EE153 pKa = 4.28NPGEE157 pKa = 4.05IQIGNSGTGSIWHH170 pKa = 6.63IAAAAFAEE178 pKa = 4.34EE179 pKa = 4.06AGIDD183 pKa = 3.95VEE185 pKa = 4.34YY186 pKa = 11.02VPYY189 pKa = 10.69DD190 pKa = 3.56GASPAVTALMGGHH203 pKa = 6.81ISAVSVSPSEE213 pKa = 3.87VRR215 pKa = 11.84GGVEE219 pKa = 4.11SGDD222 pKa = 3.73LKK224 pKa = 11.23VLGVMGEE231 pKa = 4.07EE232 pKa = 4.58RR233 pKa = 11.84DD234 pKa = 3.84PEE236 pKa = 4.7LPDD239 pKa = 3.3VPTMQEE245 pKa = 2.98AGYY248 pKa = 9.72DD249 pKa = 3.54VSVAGWGGFVAPEE262 pKa = 4.02GTPEE266 pKa = 3.99EE267 pKa = 4.16VLEE270 pKa = 4.16PLRR273 pKa = 11.84SSFKK277 pKa = 10.38NAAEE281 pKa = 4.08SEE283 pKa = 4.32EE284 pKa = 4.22FQQLMDD290 pKa = 4.45DD291 pKa = 3.75RR292 pKa = 11.84GMIPAYY298 pKa = 9.96KK299 pKa = 9.89NGKK302 pKa = 8.57EE303 pKa = 3.85FSSYY307 pKa = 11.39AEE309 pKa = 3.67EE310 pKa = 4.22QYY312 pKa = 11.43DD313 pKa = 4.04YY314 pKa = 10.87FSEE317 pKa = 5.1LIPTIEE323 pKa = 4.13INQQ326 pKa = 3.46
Molecular weight: 34.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I4NBL4|A0A1I4NBL4_9BACI Cyclic-di-AMP phosphodiesterase OS=Salibacterium qingdaonense OX=266892 GN=SAMN04488054_11658 PE=3 SV=1
MM1 pKa = 7.66GKK3 pKa = 8.01PTFSPNNRR11 pKa = 11.84KK12 pKa = 9.29RR13 pKa = 11.84KK14 pKa = 8.42KK15 pKa = 8.43VHH17 pKa = 5.59GFRR20 pKa = 11.84EE21 pKa = 3.98RR22 pKa = 11.84MSTKK26 pKa = 10.18KK27 pKa = 10.45GRR29 pKa = 11.84QVLKK33 pKa = 9.7NRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.34GRR40 pKa = 11.84KK41 pKa = 9.01VISAA45 pKa = 4.05
MM1 pKa = 7.66GKK3 pKa = 8.01PTFSPNNRR11 pKa = 11.84KK12 pKa = 9.29RR13 pKa = 11.84KK14 pKa = 8.42KK15 pKa = 8.43VHH17 pKa = 5.59GFRR20 pKa = 11.84EE21 pKa = 3.98RR22 pKa = 11.84MSTKK26 pKa = 10.18KK27 pKa = 10.45GRR29 pKa = 11.84QVLKK33 pKa = 9.7NRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.34GRR40 pKa = 11.84KK41 pKa = 9.01VISAA45 pKa = 4.05
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1002558 |
39 |
1531 |
283.8 |
31.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.837 ± 0.043 | 0.67 ± 0.013 |
5.494 ± 0.042 | 8.263 ± 0.06 |
4.259 ± 0.039 | 7.31 ± 0.044 |
2.312 ± 0.024 | 6.386 ± 0.04 |
5.309 ± 0.039 | 9.247 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.074 ± 0.023 | 3.869 ± 0.031 |
3.882 ± 0.025 | 4.043 ± 0.031 |
4.701 ± 0.036 | 6.216 ± 0.03 |
5.653 ± 0.031 | 7.036 ± 0.035 |
1.134 ± 0.018 | 3.305 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
