
Beatrice Hill virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Tibrovirus; Beatrice Hill tibrovirus
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
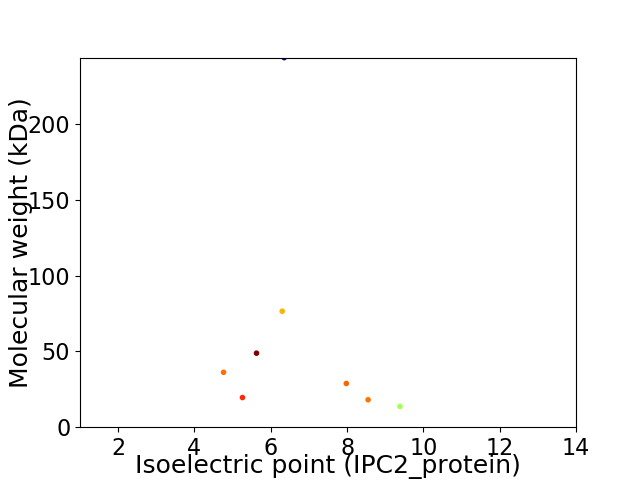
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
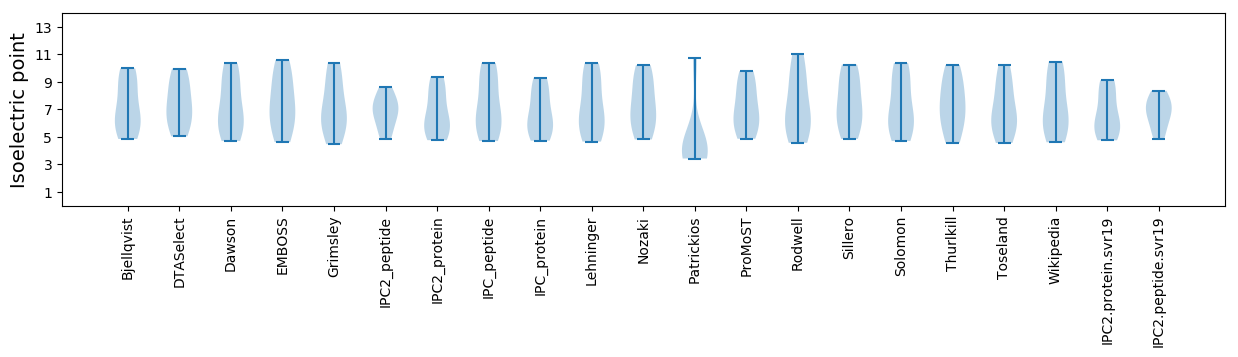
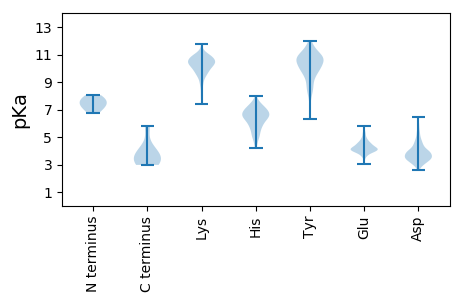
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1J0F5D8|A0A1J0F5D8_9RHAB Nucleocapsid protein OS=Beatrice Hill virus OX=1819301 GN=N PE=4 SV=1
MM1 pKa = 7.18KK2 pKa = 9.89RR3 pKa = 11.84HH4 pKa = 6.68RR5 pKa = 11.84ISIPYY10 pKa = 8.38VTDD13 pKa = 3.12QVLRR17 pKa = 11.84NTSDD21 pKa = 3.38AVDD24 pKa = 3.9PNEE27 pKa = 5.63VIDD30 pKa = 3.99QQISDD35 pKa = 5.4DD36 pKa = 3.63ITNPKK41 pKa = 9.74QDD43 pKa = 3.78LKK45 pKa = 11.25EE46 pKa = 4.04FLDD49 pKa = 3.88TRR51 pKa = 11.84EE52 pKa = 3.81LNYY55 pKa = 9.41RR56 pKa = 11.84TRR58 pKa = 11.84ASSASSYY65 pKa = 11.67DD66 pKa = 3.49DD67 pKa = 5.82DD68 pKa = 5.01DD69 pKa = 4.0WADD72 pKa = 3.85SIVDD76 pKa = 3.93LSRR79 pKa = 11.84KK80 pKa = 8.08SQHH83 pKa = 6.56KK84 pKa = 10.5SDD86 pKa = 5.55GEE88 pKa = 4.27DD89 pKa = 3.26NPQKK93 pKa = 10.97SSGGPNLVLPSISSGVQDD111 pKa = 3.26KK112 pKa = 10.97CSPNTTKK119 pKa = 10.8DD120 pKa = 3.19NKK122 pKa = 9.85IYY124 pKa = 10.82KK125 pKa = 9.5VDD127 pKa = 4.25RR128 pKa = 11.84EE129 pKa = 4.37DD130 pKa = 4.92NDD132 pKa = 3.47MGPNIHH138 pKa = 6.58QIPEE142 pKa = 3.89NRR144 pKa = 11.84PVYY147 pKa = 9.5PSQMYY152 pKa = 9.64PLLEE156 pKa = 4.1VPGNYY161 pKa = 10.05NLLIPKK167 pKa = 9.46LQFFFKK173 pKa = 10.87YY174 pKa = 9.84YY175 pKa = 11.05GLYY178 pKa = 10.24EE179 pKa = 4.36DD180 pKa = 3.76TDD182 pKa = 4.19YY183 pKa = 11.84VVDD186 pKa = 4.79KK187 pKa = 10.89DD188 pKa = 3.56NQGYY192 pKa = 8.78YY193 pKa = 10.35FYY195 pKa = 8.72PTKK198 pKa = 10.51KK199 pKa = 7.37WTTRR203 pKa = 11.84DD204 pKa = 3.2QKK206 pKa = 11.64EE207 pKa = 4.2MFEE210 pKa = 4.32CTPKK214 pKa = 10.57KK215 pKa = 10.56EE216 pKa = 4.21EE217 pKa = 4.26NEE219 pKa = 3.78DD220 pKa = 3.72DD221 pKa = 4.06ALLNLEE227 pKa = 5.57DD228 pKa = 4.86INDD231 pKa = 4.64DD232 pKa = 4.01GPFDD236 pKa = 4.56EE237 pKa = 5.82DD238 pKa = 4.39PEE240 pKa = 4.34VHH242 pKa = 6.28QLIDD246 pKa = 4.56FITKK250 pKa = 9.52GFYY253 pKa = 9.64VEE255 pKa = 4.03KK256 pKa = 10.59RR257 pKa = 11.84NIKK260 pKa = 7.78GTYY263 pKa = 9.5YY264 pKa = 10.8FDD266 pKa = 4.05INNPSLNVNKK276 pKa = 9.93IANVDD281 pKa = 3.78CQNKK285 pKa = 8.78VLSVKK290 pKa = 10.28EE291 pKa = 4.12KK292 pKa = 10.16IDD294 pKa = 4.04MIFKK298 pKa = 10.68ASGIYY303 pKa = 9.93RR304 pKa = 11.84AMKK307 pKa = 10.51LKK309 pKa = 10.82AKK311 pKa = 8.7WW312 pKa = 3.09
MM1 pKa = 7.18KK2 pKa = 9.89RR3 pKa = 11.84HH4 pKa = 6.68RR5 pKa = 11.84ISIPYY10 pKa = 8.38VTDD13 pKa = 3.12QVLRR17 pKa = 11.84NTSDD21 pKa = 3.38AVDD24 pKa = 3.9PNEE27 pKa = 5.63VIDD30 pKa = 3.99QQISDD35 pKa = 5.4DD36 pKa = 3.63ITNPKK41 pKa = 9.74QDD43 pKa = 3.78LKK45 pKa = 11.25EE46 pKa = 4.04FLDD49 pKa = 3.88TRR51 pKa = 11.84EE52 pKa = 3.81LNYY55 pKa = 9.41RR56 pKa = 11.84TRR58 pKa = 11.84ASSASSYY65 pKa = 11.67DD66 pKa = 3.49DD67 pKa = 5.82DD68 pKa = 5.01DD69 pKa = 4.0WADD72 pKa = 3.85SIVDD76 pKa = 3.93LSRR79 pKa = 11.84KK80 pKa = 8.08SQHH83 pKa = 6.56KK84 pKa = 10.5SDD86 pKa = 5.55GEE88 pKa = 4.27DD89 pKa = 3.26NPQKK93 pKa = 10.97SSGGPNLVLPSISSGVQDD111 pKa = 3.26KK112 pKa = 10.97CSPNTTKK119 pKa = 10.8DD120 pKa = 3.19NKK122 pKa = 9.85IYY124 pKa = 10.82KK125 pKa = 9.5VDD127 pKa = 4.25RR128 pKa = 11.84EE129 pKa = 4.37DD130 pKa = 4.92NDD132 pKa = 3.47MGPNIHH138 pKa = 6.58QIPEE142 pKa = 3.89NRR144 pKa = 11.84PVYY147 pKa = 9.5PSQMYY152 pKa = 9.64PLLEE156 pKa = 4.1VPGNYY161 pKa = 10.05NLLIPKK167 pKa = 9.46LQFFFKK173 pKa = 10.87YY174 pKa = 9.84YY175 pKa = 11.05GLYY178 pKa = 10.24EE179 pKa = 4.36DD180 pKa = 3.76TDD182 pKa = 4.19YY183 pKa = 11.84VVDD186 pKa = 4.79KK187 pKa = 10.89DD188 pKa = 3.56NQGYY192 pKa = 8.78YY193 pKa = 10.35FYY195 pKa = 8.72PTKK198 pKa = 10.51KK199 pKa = 7.37WTTRR203 pKa = 11.84DD204 pKa = 3.2QKK206 pKa = 11.64EE207 pKa = 4.2MFEE210 pKa = 4.32CTPKK214 pKa = 10.57KK215 pKa = 10.56EE216 pKa = 4.21EE217 pKa = 4.26NEE219 pKa = 3.78DD220 pKa = 3.72DD221 pKa = 4.06ALLNLEE227 pKa = 5.57DD228 pKa = 4.86INDD231 pKa = 4.64DD232 pKa = 4.01GPFDD236 pKa = 4.56EE237 pKa = 5.82DD238 pKa = 4.39PEE240 pKa = 4.34VHH242 pKa = 6.28QLIDD246 pKa = 4.56FITKK250 pKa = 9.52GFYY253 pKa = 9.64VEE255 pKa = 4.03KK256 pKa = 10.59RR257 pKa = 11.84NIKK260 pKa = 7.78GTYY263 pKa = 9.5YY264 pKa = 10.8FDD266 pKa = 4.05INNPSLNVNKK276 pKa = 9.93IANVDD281 pKa = 3.78CQNKK285 pKa = 8.78VLSVKK290 pKa = 10.28EE291 pKa = 4.12KK292 pKa = 10.16IDD294 pKa = 4.04MIFKK298 pKa = 10.68ASGIYY303 pKa = 9.93RR304 pKa = 11.84AMKK307 pKa = 10.51LKK309 pKa = 10.82AKK311 pKa = 8.7WW312 pKa = 3.09
Molecular weight: 36.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1J0F5M1|A0A1J0F5M1_9RHAB U3 protein OS=Beatrice Hill virus OX=1819301 GN=U3 PE=4 SV=1
MM1 pKa = 7.08TLSMDD6 pKa = 3.64RR7 pKa = 11.84LGTEE11 pKa = 4.15IFTKK15 pKa = 9.95IYY17 pKa = 10.36FKK19 pKa = 10.62IKK21 pKa = 10.38EE22 pKa = 4.55DD23 pKa = 3.73FTGWAYY29 pKa = 11.22NLWFKK34 pKa = 10.82IKK36 pKa = 10.61AIVLVLIIILVFVVTIKK53 pKa = 10.71LFKK56 pKa = 10.26SCAYY60 pKa = 9.84VINLIKK66 pKa = 10.67NCCKK70 pKa = 10.08GIAMLRR76 pKa = 11.84HH77 pKa = 6.53KK78 pKa = 10.34GRR80 pKa = 11.84KK81 pKa = 7.85ILSRR85 pKa = 11.84YY86 pKa = 9.45RR87 pKa = 11.84SDD89 pKa = 3.85RR90 pKa = 11.84KK91 pKa = 10.47KK92 pKa = 10.84EE93 pKa = 3.96GNKK96 pKa = 10.08KK97 pKa = 9.74ILIEE101 pKa = 4.19SPIYY105 pKa = 9.54MNNFGFQSDD114 pKa = 4.01TKK116 pKa = 10.95SS117 pKa = 2.97
MM1 pKa = 7.08TLSMDD6 pKa = 3.64RR7 pKa = 11.84LGTEE11 pKa = 4.15IFTKK15 pKa = 9.95IYY17 pKa = 10.36FKK19 pKa = 10.62IKK21 pKa = 10.38EE22 pKa = 4.55DD23 pKa = 3.73FTGWAYY29 pKa = 11.22NLWFKK34 pKa = 10.82IKK36 pKa = 10.61AIVLVLIIILVFVVTIKK53 pKa = 10.71LFKK56 pKa = 10.26SCAYY60 pKa = 9.84VINLIKK66 pKa = 10.67NCCKK70 pKa = 10.08GIAMLRR76 pKa = 11.84HH77 pKa = 6.53KK78 pKa = 10.34GRR80 pKa = 11.84KK81 pKa = 7.85ILSRR85 pKa = 11.84YY86 pKa = 9.45RR87 pKa = 11.84SDD89 pKa = 3.85RR90 pKa = 11.84KK91 pKa = 10.47KK92 pKa = 10.84EE93 pKa = 3.96GNKK96 pKa = 10.08KK97 pKa = 9.74ILIEE101 pKa = 4.19SPIYY105 pKa = 9.54MNNFGFQSDD114 pKa = 4.01TKK116 pKa = 10.95SS117 pKa = 2.97
Molecular weight: 13.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4214 |
117 |
2116 |
526.8 |
60.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.346 ± 0.276 | 2.207 ± 0.256 |
6.431 ± 0.856 | 5.221 ± 0.206 |
4.438 ± 0.373 | 5.031 ± 0.285 |
2.729 ± 0.448 | 8.234 ± 0.419 |
7.309 ± 0.776 | 10.228 ± 0.978 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.302 ± 0.249 | 7.048 ± 0.296 |
3.963 ± 0.324 | 3.037 ± 0.269 |
4.034 ± 0.208 | 7.712 ± 0.53 |
5.529 ± 0.331 | 4.865 ± 0.372 |
1.614 ± 0.121 | 4.722 ± 0.189 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
