
Rhizobium sp. PDO1-076
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
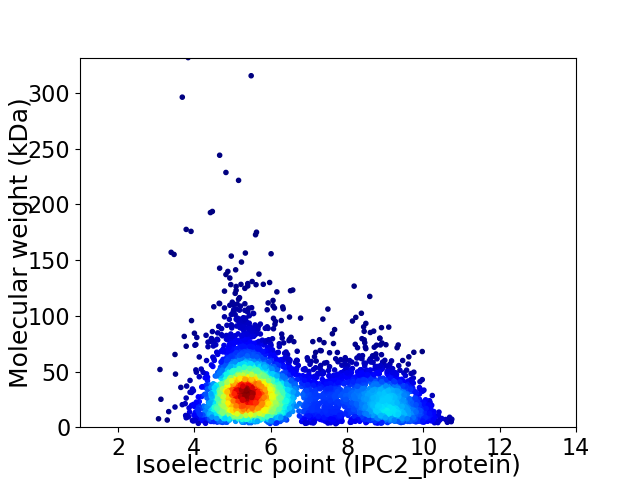
Virtual 2D-PAGE plot for 5347 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H4F4A6|H4F4A6_9RHIZ Methylglyoxal synthase OS=Rhizobium sp. PDO1-076 OX=1125979 GN=mgsA PE=3 SV=1
MM1 pKa = 7.49NIKK4 pKa = 10.39SLLLGSAAALAVVSGAQAADD24 pKa = 4.21AIVAAEE30 pKa = 4.47PEE32 pKa = 3.93PMEE35 pKa = 4.1YY36 pKa = 10.8VRR38 pKa = 11.84VCDD41 pKa = 4.42AFGTGYY47 pKa = 10.5FYY49 pKa = 10.61IPGSEE54 pKa = 4.09TCLKK58 pKa = 9.99ISGYY62 pKa = 9.93VRR64 pKa = 11.84FQTEE68 pKa = 3.85FSDD71 pKa = 5.08NGDD74 pKa = 3.64DD75 pKa = 4.54SDD77 pKa = 3.45WNMRR81 pKa = 11.84TRR83 pKa = 11.84AKK85 pKa = 10.96LNFEE89 pKa = 4.53AKK91 pKa = 10.0NDD93 pKa = 4.01SEE95 pKa = 4.78LGTIGSYY102 pKa = 9.9VAVRR106 pKa = 11.84TWADD110 pKa = 3.32NNGSDD115 pKa = 4.29TDD117 pKa = 3.65NKK119 pKa = 11.13LEE121 pKa = 4.05IDD123 pKa = 3.66EE124 pKa = 5.25AFITVGGFKK133 pKa = 10.22VGYY136 pKa = 8.78MYY138 pKa = 10.74NYY140 pKa = 9.05WDD142 pKa = 3.96NDD144 pKa = 3.44LSGEE148 pKa = 4.05TDD150 pKa = 3.98DD151 pKa = 5.36IGSNRR156 pKa = 11.84LNAIGYY162 pKa = 9.54QYY164 pKa = 10.48TADD167 pKa = 3.65AFSAGVFLDD176 pKa = 4.06EE177 pKa = 5.34LSGGYY182 pKa = 9.77SDD184 pKa = 6.14DD185 pKa = 4.34SFDD188 pKa = 3.6VAYY191 pKa = 10.59DD192 pKa = 3.56GSDD195 pKa = 3.25NLGVEE200 pKa = 4.33GQIAATFGVVTAEE213 pKa = 4.21LLGAYY218 pKa = 9.77DD219 pKa = 4.4FAAEE223 pKa = 4.05NGAIRR228 pKa = 11.84AKK230 pKa = 10.64LLADD234 pKa = 3.83VGPGTFGLAAIYY246 pKa = 8.62STGANAYY253 pKa = 9.47YY254 pKa = 10.2DD255 pKa = 3.66VSEE258 pKa = 4.08WTVAAEE264 pKa = 4.16YY265 pKa = 10.44AVKK268 pKa = 10.73LSDD271 pKa = 5.18KK272 pKa = 9.88FTLTPGVQYY281 pKa = 9.73WDD283 pKa = 3.57SYY285 pKa = 11.17GLVSADD291 pKa = 5.43DD292 pKa = 4.13FDD294 pKa = 5.21SSNDD298 pKa = 2.87AWKK301 pKa = 10.74AGLTLDD307 pKa = 3.69YY308 pKa = 10.66KK309 pKa = 10.49IAQGLTAKK317 pKa = 10.44VAVNYY322 pKa = 10.05FDD324 pKa = 5.43EE325 pKa = 5.33DD326 pKa = 3.88AADD329 pKa = 4.07DD330 pKa = 4.46GVWDD334 pKa = 3.96GFVRR338 pKa = 11.84LQRR341 pKa = 11.84SFF343 pKa = 3.42
MM1 pKa = 7.49NIKK4 pKa = 10.39SLLLGSAAALAVVSGAQAADD24 pKa = 4.21AIVAAEE30 pKa = 4.47PEE32 pKa = 3.93PMEE35 pKa = 4.1YY36 pKa = 10.8VRR38 pKa = 11.84VCDD41 pKa = 4.42AFGTGYY47 pKa = 10.5FYY49 pKa = 10.61IPGSEE54 pKa = 4.09TCLKK58 pKa = 9.99ISGYY62 pKa = 9.93VRR64 pKa = 11.84FQTEE68 pKa = 3.85FSDD71 pKa = 5.08NGDD74 pKa = 3.64DD75 pKa = 4.54SDD77 pKa = 3.45WNMRR81 pKa = 11.84TRR83 pKa = 11.84AKK85 pKa = 10.96LNFEE89 pKa = 4.53AKK91 pKa = 10.0NDD93 pKa = 4.01SEE95 pKa = 4.78LGTIGSYY102 pKa = 9.9VAVRR106 pKa = 11.84TWADD110 pKa = 3.32NNGSDD115 pKa = 4.29TDD117 pKa = 3.65NKK119 pKa = 11.13LEE121 pKa = 4.05IDD123 pKa = 3.66EE124 pKa = 5.25AFITVGGFKK133 pKa = 10.22VGYY136 pKa = 8.78MYY138 pKa = 10.74NYY140 pKa = 9.05WDD142 pKa = 3.96NDD144 pKa = 3.44LSGEE148 pKa = 4.05TDD150 pKa = 3.98DD151 pKa = 5.36IGSNRR156 pKa = 11.84LNAIGYY162 pKa = 9.54QYY164 pKa = 10.48TADD167 pKa = 3.65AFSAGVFLDD176 pKa = 4.06EE177 pKa = 5.34LSGGYY182 pKa = 9.77SDD184 pKa = 6.14DD185 pKa = 4.34SFDD188 pKa = 3.6VAYY191 pKa = 10.59DD192 pKa = 3.56GSDD195 pKa = 3.25NLGVEE200 pKa = 4.33GQIAATFGVVTAEE213 pKa = 4.21LLGAYY218 pKa = 9.77DD219 pKa = 4.4FAAEE223 pKa = 4.05NGAIRR228 pKa = 11.84AKK230 pKa = 10.64LLADD234 pKa = 3.83VGPGTFGLAAIYY246 pKa = 8.62STGANAYY253 pKa = 9.47YY254 pKa = 10.2DD255 pKa = 3.66VSEE258 pKa = 4.08WTVAAEE264 pKa = 4.16YY265 pKa = 10.44AVKK268 pKa = 10.73LSDD271 pKa = 5.18KK272 pKa = 9.88FTLTPGVQYY281 pKa = 9.73WDD283 pKa = 3.57SYY285 pKa = 11.17GLVSADD291 pKa = 5.43DD292 pKa = 4.13FDD294 pKa = 5.21SSNDD298 pKa = 2.87AWKK301 pKa = 10.74AGLTLDD307 pKa = 3.69YY308 pKa = 10.66KK309 pKa = 10.49IAQGLTAKK317 pKa = 10.44VAVNYY322 pKa = 10.05FDD324 pKa = 5.43EE325 pKa = 5.33DD326 pKa = 3.88AADD329 pKa = 4.07DD330 pKa = 4.46GVWDD334 pKa = 3.96GFVRR338 pKa = 11.84LQRR341 pKa = 11.84SFF343 pKa = 3.42
Molecular weight: 36.92 kDa
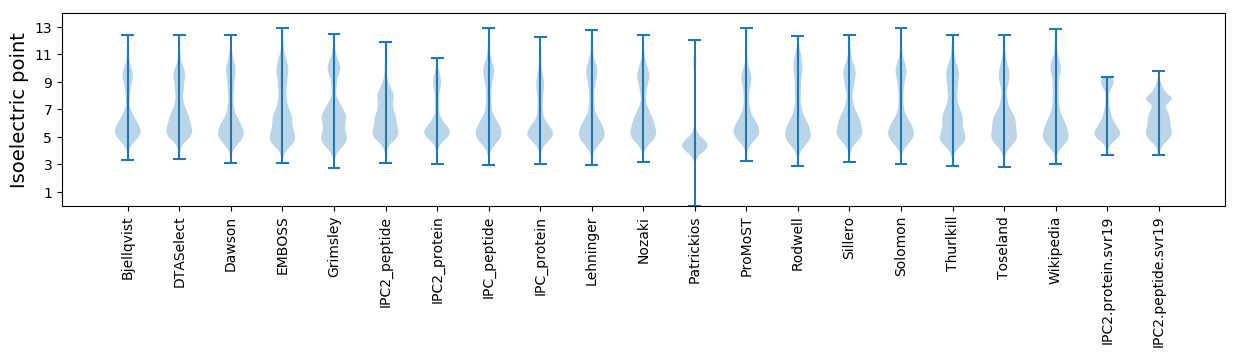
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H4F1F9|H4F1F9_9RHIZ Type IV secretion system protein virB4 OS=Rhizobium sp. PDO1-076 OX=1125979 GN=PDO_4199 PE=3 SV=1
MM1 pKa = 7.83TDD3 pKa = 3.13TPTNNANGPASAMPVVVMTAGGLNPTLVIQALAARR38 pKa = 11.84NADD41 pKa = 2.62IHH43 pKa = 7.15VILEE47 pKa = 4.15QPEE50 pKa = 4.35SKK52 pKa = 10.86LDD54 pKa = 3.22ITRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84ARR61 pKa = 11.84RR62 pKa = 11.84LGWVAAIGQLATMIAARR79 pKa = 11.84LLRR82 pKa = 11.84KK83 pKa = 9.19LANKK87 pKa = 10.22RR88 pKa = 11.84IRR90 pKa = 11.84QILDD94 pKa = 2.96AHH96 pKa = 6.32NLTDD100 pKa = 3.63TMPGNVAVHH109 pKa = 5.56QVTSINDD116 pKa = 3.24QEE118 pKa = 4.25TRR120 pKa = 11.84ALIAQIRR127 pKa = 11.84PAAILLVSTRR137 pKa = 11.84LMARR141 pKa = 11.84KK142 pKa = 8.89QLAAMPCPVLNLHH155 pKa = 7.03AGINPAYY162 pKa = 8.78RR163 pKa = 11.84GQMGGYY169 pKa = 8.31WSRR172 pKa = 11.84RR173 pKa = 11.84LNDD176 pKa = 3.14ACNFGATVHH185 pKa = 6.35LVDD188 pKa = 5.34AGTDD192 pKa = 3.37TGGTLYY198 pKa = 10.15QVRR201 pKa = 11.84TTPAAGDD208 pKa = 4.34FISTYY213 pKa = 10.6PLVLTVAALDD223 pKa = 3.53ITCRR227 pKa = 11.84AVADD231 pKa = 4.0ALASNLQPIAPKK243 pKa = 10.55GPSALHH249 pKa = 6.2FPPTLWSWIFHH260 pKa = 6.58GITRR264 pKa = 11.84RR265 pKa = 11.84IWW267 pKa = 3.08
MM1 pKa = 7.83TDD3 pKa = 3.13TPTNNANGPASAMPVVVMTAGGLNPTLVIQALAARR38 pKa = 11.84NADD41 pKa = 2.62IHH43 pKa = 7.15VILEE47 pKa = 4.15QPEE50 pKa = 4.35SKK52 pKa = 10.86LDD54 pKa = 3.22ITRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84ARR61 pKa = 11.84RR62 pKa = 11.84LGWVAAIGQLATMIAARR79 pKa = 11.84LLRR82 pKa = 11.84KK83 pKa = 9.19LANKK87 pKa = 10.22RR88 pKa = 11.84IRR90 pKa = 11.84QILDD94 pKa = 2.96AHH96 pKa = 6.32NLTDD100 pKa = 3.63TMPGNVAVHH109 pKa = 5.56QVTSINDD116 pKa = 3.24QEE118 pKa = 4.25TRR120 pKa = 11.84ALIAQIRR127 pKa = 11.84PAAILLVSTRR137 pKa = 11.84LMARR141 pKa = 11.84KK142 pKa = 8.89QLAAMPCPVLNLHH155 pKa = 7.03AGINPAYY162 pKa = 8.78RR163 pKa = 11.84GQMGGYY169 pKa = 8.31WSRR172 pKa = 11.84RR173 pKa = 11.84LNDD176 pKa = 3.14ACNFGATVHH185 pKa = 6.35LVDD188 pKa = 5.34AGTDD192 pKa = 3.37TGGTLYY198 pKa = 10.15QVRR201 pKa = 11.84TTPAAGDD208 pKa = 4.34FISTYY213 pKa = 10.6PLVLTVAALDD223 pKa = 3.53ITCRR227 pKa = 11.84AVADD231 pKa = 4.0ALASNLQPIAPKK243 pKa = 10.55GPSALHH249 pKa = 6.2FPPTLWSWIFHH260 pKa = 6.58GITRR264 pKa = 11.84RR265 pKa = 11.84IWW267 pKa = 3.08
Molecular weight: 28.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1610893 |
30 |
3141 |
301.3 |
32.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.808 ± 0.044 | 0.801 ± 0.01 |
5.842 ± 0.029 | 5.7 ± 0.029 |
3.866 ± 0.024 | 8.258 ± 0.034 |
2.047 ± 0.016 | 5.757 ± 0.024 |
3.646 ± 0.03 | 10.065 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.609 ± 0.017 | 2.846 ± 0.025 |
4.767 ± 0.026 | 3.203 ± 0.023 |
6.554 ± 0.038 | 5.902 ± 0.033 |
5.494 ± 0.035 | 7.327 ± 0.027 |
1.237 ± 0.014 | 2.27 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
