
Lake Sarah-associated circular virus-5
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
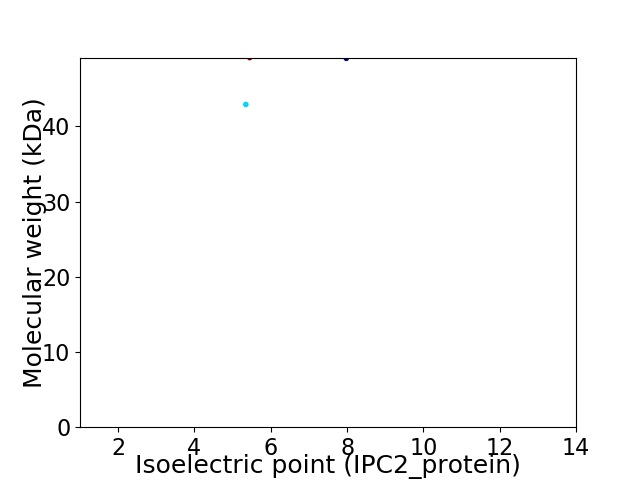
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GA52|A0A126GA52_9VIRU Coat protein OS=Lake Sarah-associated circular virus-5 OX=1685779 PE=4 SV=1
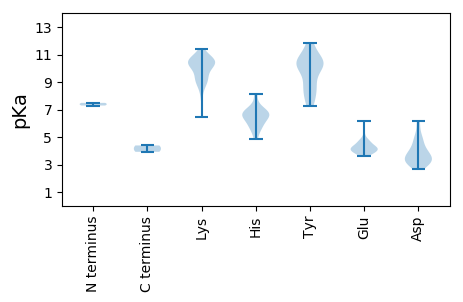
MM1 pKa = 7.29STRR4 pKa = 11.84QRR6 pKa = 11.84HH7 pKa = 4.87RR8 pKa = 11.84SVCFTLNNYY17 pKa = 7.39TAQEE21 pKa = 4.05KK22 pKa = 7.89QHH24 pKa = 6.47IKK26 pKa = 10.74DD27 pKa = 3.47GADD30 pKa = 2.94AGSFKK35 pKa = 11.11YY36 pKa = 10.28IVFQEE41 pKa = 3.73EE42 pKa = 4.02RR43 pKa = 11.84GAVGTPHH50 pKa = 6.32LQGYY54 pKa = 7.26AQRR57 pKa = 11.84AQPTEE62 pKa = 3.77FKK64 pKa = 8.69TWKK67 pKa = 9.98RR68 pKa = 11.84FLGDD72 pKa = 3.26RR73 pKa = 11.84CHH75 pKa = 8.13LEE77 pKa = 4.13GSKK80 pKa = 9.29GTPARR85 pKa = 11.84NRR87 pKa = 11.84EE88 pKa = 4.26YY89 pKa = 9.96CTKK92 pKa = 10.6DD93 pKa = 2.87ADD95 pKa = 4.96RR96 pKa = 11.84IPGTLYY102 pKa = 10.39FEE104 pKa = 4.68KK105 pKa = 11.11GEE107 pKa = 4.18VPVQGEE113 pKa = 4.46RR114 pKa = 11.84NDD116 pKa = 3.34LTAIFEE122 pKa = 4.14AAKK125 pKa = 10.51DD126 pKa = 3.66STVSVRR132 pKa = 11.84DD133 pKa = 3.6LVDD136 pKa = 3.21TNGAAFIRR144 pKa = 11.84YY145 pKa = 8.26YY146 pKa = 10.81KK147 pKa = 10.63GVLAVRR153 pKa = 11.84SIFSDD158 pKa = 2.72KK159 pKa = 10.41RR160 pKa = 11.84QRR162 pKa = 11.84KK163 pKa = 6.48TKK165 pKa = 10.1VFWFYY170 pKa = 11.63GPTGLGKK177 pKa = 9.74SHH179 pKa = 6.65EE180 pKa = 4.77CNRR183 pKa = 11.84LAPNAFWKK191 pKa = 10.49QNSPWWCGYY200 pKa = 9.78DD201 pKa = 4.03ANEE204 pKa = 4.86HH205 pKa = 6.69DD206 pKa = 6.14DD207 pKa = 4.64IVIDD211 pKa = 4.51DD212 pKa = 4.04YY213 pKa = 11.73RR214 pKa = 11.84ADD216 pKa = 3.52FCKK219 pKa = 10.22FAQLLRR225 pKa = 11.84LFDD228 pKa = 3.85FTKK231 pKa = 10.08LTVEE235 pKa = 4.35TKK237 pKa = 10.39GGNVNFRR244 pKa = 11.84AKK246 pKa = 10.25RR247 pKa = 11.84IFISTPRR254 pKa = 11.84SPLEE258 pKa = 3.61TWIGRR263 pKa = 11.84SEE265 pKa = 4.2EE266 pKa = 5.22DD267 pKa = 2.77IQQLLRR273 pKa = 11.84RR274 pKa = 11.84IEE276 pKa = 4.04VLVEE280 pKa = 4.15FTPSPFTDD288 pKa = 3.19NGRR291 pKa = 11.84VLPNKK296 pKa = 9.23VFHH299 pKa = 6.81KK300 pKa = 10.6GSPDD304 pKa = 3.61DD305 pKa = 5.28LVVDD309 pKa = 4.54DD310 pKa = 6.13LGDD313 pKa = 4.02GDD315 pKa = 3.98QQPRR319 pKa = 11.84SPVEE323 pKa = 4.03TPPEE327 pKa = 3.93EE328 pKa = 4.24AVEE331 pKa = 4.25DD332 pKa = 3.97ASDD335 pKa = 3.13RR336 pKa = 11.84RR337 pKa = 11.84TRR339 pKa = 11.84PRR341 pKa = 11.84VEE343 pKa = 4.01EE344 pKa = 3.79ADD346 pKa = 3.67EE347 pKa = 4.2PRR349 pKa = 11.84EE350 pKa = 3.78QFAQDD355 pKa = 3.02EE356 pKa = 4.72YY357 pKa = 11.83GFLNEE362 pKa = 4.76FDD364 pKa = 4.71EE365 pKa = 6.17LNDD368 pKa = 4.85DD369 pKa = 5.03DD370 pKa = 4.8FTFF373 pKa = 4.39
MM1 pKa = 7.29STRR4 pKa = 11.84QRR6 pKa = 11.84HH7 pKa = 4.87RR8 pKa = 11.84SVCFTLNNYY17 pKa = 7.39TAQEE21 pKa = 4.05KK22 pKa = 7.89QHH24 pKa = 6.47IKK26 pKa = 10.74DD27 pKa = 3.47GADD30 pKa = 2.94AGSFKK35 pKa = 11.11YY36 pKa = 10.28IVFQEE41 pKa = 3.73EE42 pKa = 4.02RR43 pKa = 11.84GAVGTPHH50 pKa = 6.32LQGYY54 pKa = 7.26AQRR57 pKa = 11.84AQPTEE62 pKa = 3.77FKK64 pKa = 8.69TWKK67 pKa = 9.98RR68 pKa = 11.84FLGDD72 pKa = 3.26RR73 pKa = 11.84CHH75 pKa = 8.13LEE77 pKa = 4.13GSKK80 pKa = 9.29GTPARR85 pKa = 11.84NRR87 pKa = 11.84EE88 pKa = 4.26YY89 pKa = 9.96CTKK92 pKa = 10.6DD93 pKa = 2.87ADD95 pKa = 4.96RR96 pKa = 11.84IPGTLYY102 pKa = 10.39FEE104 pKa = 4.68KK105 pKa = 11.11GEE107 pKa = 4.18VPVQGEE113 pKa = 4.46RR114 pKa = 11.84NDD116 pKa = 3.34LTAIFEE122 pKa = 4.14AAKK125 pKa = 10.51DD126 pKa = 3.66STVSVRR132 pKa = 11.84DD133 pKa = 3.6LVDD136 pKa = 3.21TNGAAFIRR144 pKa = 11.84YY145 pKa = 8.26YY146 pKa = 10.81KK147 pKa = 10.63GVLAVRR153 pKa = 11.84SIFSDD158 pKa = 2.72KK159 pKa = 10.41RR160 pKa = 11.84QRR162 pKa = 11.84KK163 pKa = 6.48TKK165 pKa = 10.1VFWFYY170 pKa = 11.63GPTGLGKK177 pKa = 9.74SHH179 pKa = 6.65EE180 pKa = 4.77CNRR183 pKa = 11.84LAPNAFWKK191 pKa = 10.49QNSPWWCGYY200 pKa = 9.78DD201 pKa = 4.03ANEE204 pKa = 4.86HH205 pKa = 6.69DD206 pKa = 6.14DD207 pKa = 4.64IVIDD211 pKa = 4.51DD212 pKa = 4.04YY213 pKa = 11.73RR214 pKa = 11.84ADD216 pKa = 3.52FCKK219 pKa = 10.22FAQLLRR225 pKa = 11.84LFDD228 pKa = 3.85FTKK231 pKa = 10.08LTVEE235 pKa = 4.35TKK237 pKa = 10.39GGNVNFRR244 pKa = 11.84AKK246 pKa = 10.25RR247 pKa = 11.84IFISTPRR254 pKa = 11.84SPLEE258 pKa = 3.61TWIGRR263 pKa = 11.84SEE265 pKa = 4.2EE266 pKa = 5.22DD267 pKa = 2.77IQQLLRR273 pKa = 11.84RR274 pKa = 11.84IEE276 pKa = 4.04VLVEE280 pKa = 4.15FTPSPFTDD288 pKa = 3.19NGRR291 pKa = 11.84VLPNKK296 pKa = 9.23VFHH299 pKa = 6.81KK300 pKa = 10.6GSPDD304 pKa = 3.61DD305 pKa = 5.28LVVDD309 pKa = 4.54DD310 pKa = 6.13LGDD313 pKa = 4.02GDD315 pKa = 3.98QQPRR319 pKa = 11.84SPVEE323 pKa = 4.03TPPEE327 pKa = 3.93EE328 pKa = 4.24AVEE331 pKa = 4.25DD332 pKa = 3.97ASDD335 pKa = 3.13RR336 pKa = 11.84RR337 pKa = 11.84TRR339 pKa = 11.84PRR341 pKa = 11.84VEE343 pKa = 4.01EE344 pKa = 3.79ADD346 pKa = 3.67EE347 pKa = 4.2PRR349 pKa = 11.84EE350 pKa = 3.78QFAQDD355 pKa = 3.02EE356 pKa = 4.72YY357 pKa = 11.83GFLNEE362 pKa = 4.76FDD364 pKa = 4.71EE365 pKa = 6.17LNDD368 pKa = 4.85DD369 pKa = 5.03DD370 pKa = 4.8FTFF373 pKa = 4.39
Molecular weight: 42.92 kDa
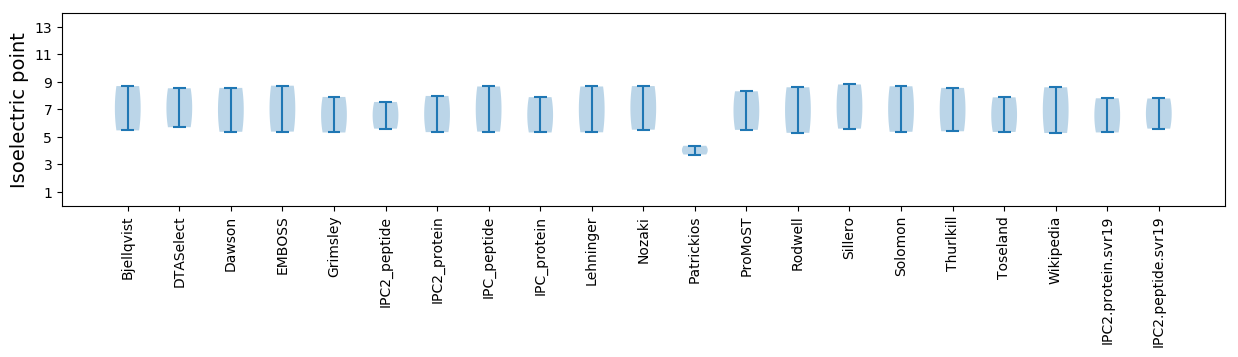
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GA52|A0A126GA52_9VIRU Coat protein OS=Lake Sarah-associated circular virus-5 OX=1685779 PE=4 SV=1
MM1 pKa = 7.49MSMSMYY7 pKa = 10.25SGSGGGKK14 pKa = 9.03SSYY17 pKa = 10.37SINYY21 pKa = 7.48SSKK24 pKa = 10.19RR25 pKa = 11.84GRR27 pKa = 11.84SPTPYY32 pKa = 10.51SSFKK36 pKa = 9.9SARR39 pKa = 11.84VQPVNQTTASAINMEE54 pKa = 4.42HH55 pKa = 7.2LNRR58 pKa = 11.84TEE60 pKa = 3.81IYY62 pKa = 10.29LGRR65 pKa = 11.84TVATASGSKK74 pKa = 10.39AYY76 pKa = 10.26GGSKK80 pKa = 9.93ANKK83 pKa = 9.29GKK85 pKa = 10.53GSKK88 pKa = 9.48ISYY91 pKa = 8.67DD92 pKa = 3.2AYY94 pKa = 10.53MNTLFPPYY102 pKa = 8.79SQTVIGYY109 pKa = 9.5GNNYY113 pKa = 8.9YY114 pKa = 11.03NKK116 pKa = 9.24TKK118 pKa = 10.66NATNSASAGDD128 pKa = 4.15DD129 pKa = 3.05KK130 pKa = 11.4SIYY133 pKa = 9.23IAQGRR138 pKa = 11.84QAWRR142 pKa = 11.84EE143 pKa = 3.86YY144 pKa = 10.66VAMPFQRR151 pKa = 11.84TAGTNGWGDD160 pKa = 4.2LFTTSVTDD168 pKa = 4.3LLVKK172 pKa = 10.6AQDD175 pKa = 3.67VLTTAVAQSVYY186 pKa = 8.15TTTTLVPSSTTTVAEE201 pKa = 4.07RR202 pKa = 11.84QKK204 pKa = 10.74LQNLSFHH211 pKa = 6.52YY212 pKa = 10.06SGGYY216 pKa = 7.73QRR218 pKa = 11.84HH219 pKa = 5.7KK220 pKa = 10.63FVNTGNSKK228 pKa = 8.44ITIEE232 pKa = 4.04FFEE235 pKa = 4.48AKK237 pKa = 9.34PRR239 pKa = 11.84NPMPAYY245 pKa = 9.93GSSATYY251 pKa = 8.78TVNSIGASIVQDD263 pKa = 3.3MQTNVPRR270 pKa = 11.84DD271 pKa = 3.39NSFYY275 pKa = 10.4PSYY278 pKa = 10.57DD279 pKa = 3.06TTDD282 pKa = 3.55FDD284 pKa = 5.4NINDD288 pKa = 3.49QLVRR292 pKa = 11.84LNAQCATTNYY302 pKa = 9.97KK303 pKa = 8.86WLCNKK308 pKa = 8.91PVKK311 pKa = 10.4HH312 pKa = 5.8VVDD315 pKa = 4.03PGGEE319 pKa = 4.3FIYY322 pKa = 10.64TMGIDD327 pKa = 2.96AFEE330 pKa = 4.39FKK332 pKa = 10.58EE333 pKa = 4.63SEE335 pKa = 4.29FNTLPGLTSTDD346 pKa = 2.71ISGVQWFYY354 pKa = 11.31PKK356 pKa = 9.06FTKK359 pKa = 10.48FLVCRR364 pKa = 11.84FTTEE368 pKa = 3.95IGHH371 pKa = 5.58STDD374 pKa = 3.2GVSGFDD380 pKa = 3.12QTYY383 pKa = 8.16TGVGYY388 pKa = 10.66VEE390 pKa = 4.61GMMAHH395 pKa = 6.72TCTEE399 pKa = 3.83HH400 pKa = 6.09HH401 pKa = 6.81NCRR404 pKa = 11.84FMPIIYY410 pKa = 8.37THH412 pKa = 6.81SNVTNNRR419 pKa = 11.84LDD421 pKa = 3.69KK422 pKa = 10.18DD423 pKa = 3.54TAAIKK428 pKa = 10.7VVFNDD433 pKa = 3.3ATGAPTNVMDD443 pKa = 5.81DD444 pKa = 3.47VV445 pKa = 3.93
MM1 pKa = 7.49MSMSMYY7 pKa = 10.25SGSGGGKK14 pKa = 9.03SSYY17 pKa = 10.37SINYY21 pKa = 7.48SSKK24 pKa = 10.19RR25 pKa = 11.84GRR27 pKa = 11.84SPTPYY32 pKa = 10.51SSFKK36 pKa = 9.9SARR39 pKa = 11.84VQPVNQTTASAINMEE54 pKa = 4.42HH55 pKa = 7.2LNRR58 pKa = 11.84TEE60 pKa = 3.81IYY62 pKa = 10.29LGRR65 pKa = 11.84TVATASGSKK74 pKa = 10.39AYY76 pKa = 10.26GGSKK80 pKa = 9.93ANKK83 pKa = 9.29GKK85 pKa = 10.53GSKK88 pKa = 9.48ISYY91 pKa = 8.67DD92 pKa = 3.2AYY94 pKa = 10.53MNTLFPPYY102 pKa = 8.79SQTVIGYY109 pKa = 9.5GNNYY113 pKa = 8.9YY114 pKa = 11.03NKK116 pKa = 9.24TKK118 pKa = 10.66NATNSASAGDD128 pKa = 4.15DD129 pKa = 3.05KK130 pKa = 11.4SIYY133 pKa = 9.23IAQGRR138 pKa = 11.84QAWRR142 pKa = 11.84EE143 pKa = 3.86YY144 pKa = 10.66VAMPFQRR151 pKa = 11.84TAGTNGWGDD160 pKa = 4.2LFTTSVTDD168 pKa = 4.3LLVKK172 pKa = 10.6AQDD175 pKa = 3.67VLTTAVAQSVYY186 pKa = 8.15TTTTLVPSSTTTVAEE201 pKa = 4.07RR202 pKa = 11.84QKK204 pKa = 10.74LQNLSFHH211 pKa = 6.52YY212 pKa = 10.06SGGYY216 pKa = 7.73QRR218 pKa = 11.84HH219 pKa = 5.7KK220 pKa = 10.63FVNTGNSKK228 pKa = 8.44ITIEE232 pKa = 4.04FFEE235 pKa = 4.48AKK237 pKa = 9.34PRR239 pKa = 11.84NPMPAYY245 pKa = 9.93GSSATYY251 pKa = 8.78TVNSIGASIVQDD263 pKa = 3.3MQTNVPRR270 pKa = 11.84DD271 pKa = 3.39NSFYY275 pKa = 10.4PSYY278 pKa = 10.57DD279 pKa = 3.06TTDD282 pKa = 3.55FDD284 pKa = 5.4NINDD288 pKa = 3.49QLVRR292 pKa = 11.84LNAQCATTNYY302 pKa = 9.97KK303 pKa = 8.86WLCNKK308 pKa = 8.91PVKK311 pKa = 10.4HH312 pKa = 5.8VVDD315 pKa = 4.03PGGEE319 pKa = 4.3FIYY322 pKa = 10.64TMGIDD327 pKa = 2.96AFEE330 pKa = 4.39FKK332 pKa = 10.58EE333 pKa = 4.63SEE335 pKa = 4.29FNTLPGLTSTDD346 pKa = 2.71ISGVQWFYY354 pKa = 11.31PKK356 pKa = 9.06FTKK359 pKa = 10.48FLVCRR364 pKa = 11.84FTTEE368 pKa = 3.95IGHH371 pKa = 5.58STDD374 pKa = 3.2GVSGFDD380 pKa = 3.12QTYY383 pKa = 8.16TGVGYY388 pKa = 10.66VEE390 pKa = 4.61GMMAHH395 pKa = 6.72TCTEE399 pKa = 3.83HH400 pKa = 6.09HH401 pKa = 6.81NCRR404 pKa = 11.84FMPIIYY410 pKa = 8.37THH412 pKa = 6.81SNVTNNRR419 pKa = 11.84LDD421 pKa = 3.69KK422 pKa = 10.18DD423 pKa = 3.54TAAIKK428 pKa = 10.7VVFNDD433 pKa = 3.3ATGAPTNVMDD443 pKa = 5.81DD444 pKa = 3.47VV445 pKa = 3.93
Molecular weight: 49.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
818 |
373 |
445 |
409.0 |
45.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.724 ± 0.181 | 1.345 ± 0.165 |
6.724 ± 1.33 | 5.134 ± 1.653 |
5.746 ± 0.767 | 7.335 ± 0.396 |
1.956 ± 0.05 | 3.912 ± 0.267 |
5.501 ± 0.081 | 4.89 ± 0.799 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.834 ± 0.98 | 5.623 ± 1.003 |
4.645 ± 0.449 | 4.156 ± 0.251 |
5.868 ± 1.698 | 7.213 ± 1.662 |
9.046 ± 1.636 | 6.479 ± 0.196 |
1.222 ± 0.242 | 4.645 ± 1.062 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
