
Alysiella filiformis DSM 16848
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Neisseriales; Neisseriaceae; Alysiella; Alysiella filiformis
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
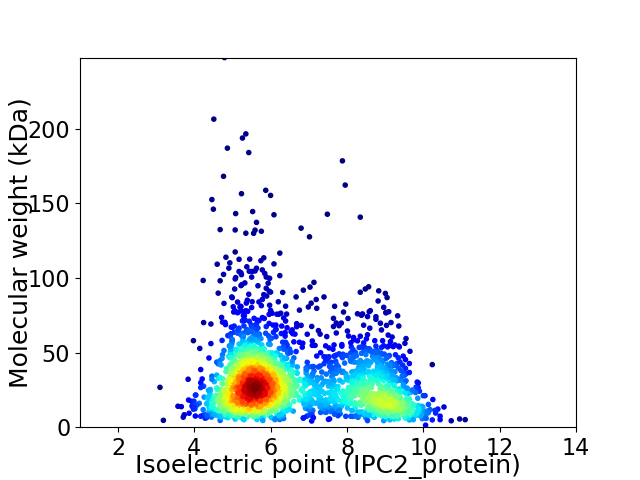
Virtual 2D-PAGE plot for 2283 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
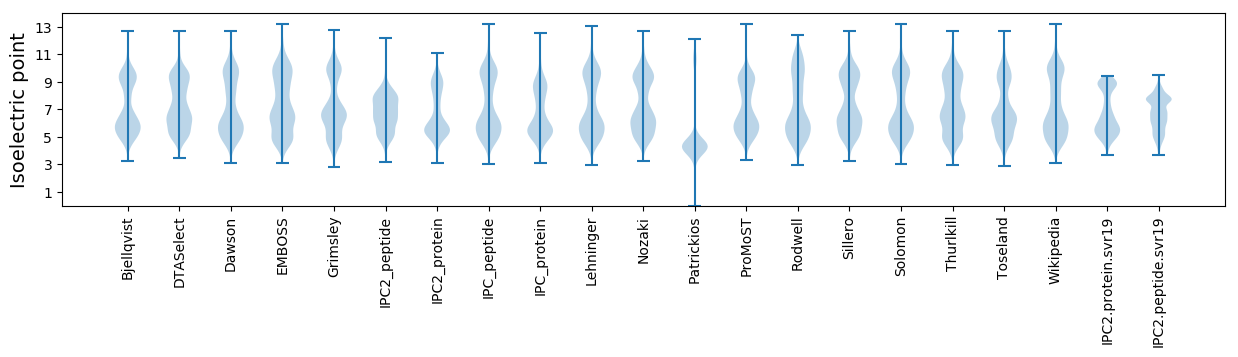
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A286E1U2|A0A286E1U2_9NEIS Histidine kinase OS=Alysiella filiformis DSM 16848 OX=1120981 GN=SAMN02746062_00071 PE=4 SV=1
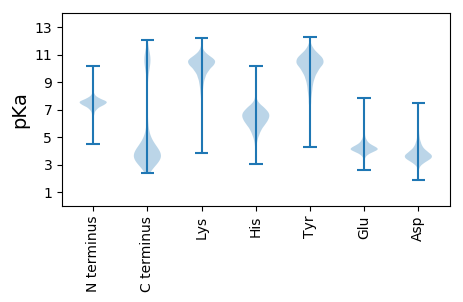
MM1 pKa = 7.44SLNIPVEE8 pKa = 4.2FQDD11 pKa = 4.4FADD14 pKa = 5.39EE15 pKa = 5.16IIASCKK21 pKa = 9.47PSVPFTLTVAPTQLWDD37 pKa = 3.71SKK39 pKa = 11.56VGGKK43 pKa = 9.63PYY45 pKa = 10.51LPRR48 pKa = 11.84DD49 pKa = 3.21FAYY52 pKa = 9.98PNNPDD57 pKa = 3.54GEE59 pKa = 4.47PLFFLAQINFAQMSALPDD77 pKa = 4.11FPSKK81 pKa = 10.98GILQFFIDD89 pKa = 5.21GKK91 pKa = 10.32DD92 pKa = 3.07DD93 pKa = 3.84CYY95 pKa = 11.38GINFDD100 pKa = 4.12NYY102 pKa = 10.53QDD104 pKa = 3.51NSHH107 pKa = 6.41YY108 pKa = 10.89KK109 pKa = 10.37LIYY112 pKa = 9.54HH113 pKa = 7.02ADD115 pKa = 3.39VVEE118 pKa = 4.66NADD121 pKa = 4.21EE122 pKa = 4.24LQQDD126 pKa = 3.65FDD128 pKa = 4.27FVQFDD133 pKa = 3.8EE134 pKa = 4.69TDD136 pKa = 3.26SGVPFPPNHH145 pKa = 6.67SYY147 pKa = 10.82QIYY150 pKa = 8.9FQAATPQPITQHH162 pKa = 6.71DD163 pKa = 3.96FAFGEE168 pKa = 4.53VVPEE172 pKa = 4.23LFDD175 pKa = 4.4FLDD178 pKa = 4.32EE179 pKa = 4.67KK180 pKa = 11.47SEE182 pKa = 4.48DD183 pKa = 5.32DD184 pKa = 3.89DD185 pKa = 5.35YY186 pKa = 12.13EE187 pKa = 4.09WDD189 pKa = 3.51LHH191 pKa = 6.74DD192 pKa = 6.0KK193 pKa = 10.95YY194 pKa = 11.6DD195 pKa = 3.65EE196 pKa = 4.43FSNSEE201 pKa = 3.61QHH203 pKa = 6.61RR204 pKa = 11.84VGGYY208 pKa = 9.83PYY210 pKa = 9.35FTQDD214 pKa = 3.09DD215 pKa = 3.85VRR217 pKa = 11.84GYY219 pKa = 11.34GSYY222 pKa = 10.8DD223 pKa = 3.17LKK225 pKa = 11.34DD226 pKa = 3.67YY227 pKa = 11.45VLLFQLVSDD236 pKa = 4.09WRR238 pKa = 11.84DD239 pKa = 3.18SNDD242 pKa = 3.99DD243 pKa = 3.78VNIMWGDD250 pKa = 3.6CGVGNFFIRR259 pKa = 11.84PEE261 pKa = 4.0DD262 pKa = 3.78LKK264 pKa = 11.57NLAFDD269 pKa = 3.79KK270 pKa = 11.31AFFTMDD276 pKa = 3.35CCC278 pKa = 5.69
MM1 pKa = 7.44SLNIPVEE8 pKa = 4.2FQDD11 pKa = 4.4FADD14 pKa = 5.39EE15 pKa = 5.16IIASCKK21 pKa = 9.47PSVPFTLTVAPTQLWDD37 pKa = 3.71SKK39 pKa = 11.56VGGKK43 pKa = 9.63PYY45 pKa = 10.51LPRR48 pKa = 11.84DD49 pKa = 3.21FAYY52 pKa = 9.98PNNPDD57 pKa = 3.54GEE59 pKa = 4.47PLFFLAQINFAQMSALPDD77 pKa = 4.11FPSKK81 pKa = 10.98GILQFFIDD89 pKa = 5.21GKK91 pKa = 10.32DD92 pKa = 3.07DD93 pKa = 3.84CYY95 pKa = 11.38GINFDD100 pKa = 4.12NYY102 pKa = 10.53QDD104 pKa = 3.51NSHH107 pKa = 6.41YY108 pKa = 10.89KK109 pKa = 10.37LIYY112 pKa = 9.54HH113 pKa = 7.02ADD115 pKa = 3.39VVEE118 pKa = 4.66NADD121 pKa = 4.21EE122 pKa = 4.24LQQDD126 pKa = 3.65FDD128 pKa = 4.27FVQFDD133 pKa = 3.8EE134 pKa = 4.69TDD136 pKa = 3.26SGVPFPPNHH145 pKa = 6.67SYY147 pKa = 10.82QIYY150 pKa = 8.9FQAATPQPITQHH162 pKa = 6.71DD163 pKa = 3.96FAFGEE168 pKa = 4.53VVPEE172 pKa = 4.23LFDD175 pKa = 4.4FLDD178 pKa = 4.32EE179 pKa = 4.67KK180 pKa = 11.47SEE182 pKa = 4.48DD183 pKa = 5.32DD184 pKa = 3.89DD185 pKa = 5.35YY186 pKa = 12.13EE187 pKa = 4.09WDD189 pKa = 3.51LHH191 pKa = 6.74DD192 pKa = 6.0KK193 pKa = 10.95YY194 pKa = 11.6DD195 pKa = 3.65EE196 pKa = 4.43FSNSEE201 pKa = 3.61QHH203 pKa = 6.61RR204 pKa = 11.84VGGYY208 pKa = 9.83PYY210 pKa = 9.35FTQDD214 pKa = 3.09DD215 pKa = 3.85VRR217 pKa = 11.84GYY219 pKa = 11.34GSYY222 pKa = 10.8DD223 pKa = 3.17LKK225 pKa = 11.34DD226 pKa = 3.67YY227 pKa = 11.45VLLFQLVSDD236 pKa = 4.09WRR238 pKa = 11.84DD239 pKa = 3.18SNDD242 pKa = 3.99DD243 pKa = 3.78VNIMWGDD250 pKa = 3.6CGVGNFFIRR259 pKa = 11.84PEE261 pKa = 4.0DD262 pKa = 3.78LKK264 pKa = 11.57NLAFDD269 pKa = 3.79KK270 pKa = 11.31AFFTMDD276 pKa = 3.35CCC278 pKa = 5.69
Molecular weight: 32.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A286EAI6|A0A286EAI6_9NEIS GTPase Era OS=Alysiella filiformis DSM 16848 OX=1120981 GN=era PE=3 SV=1
MM1 pKa = 7.63ALRR4 pKa = 11.84RR5 pKa = 11.84HH6 pKa = 4.7THH8 pKa = 7.05ALGLWHH14 pKa = 6.39ATVRR18 pKa = 11.84ALKK21 pKa = 10.4IGFQNKK27 pKa = 8.82NMLFAKK33 pKa = 9.17PRR35 pKa = 11.84LMFRR39 pKa = 11.84QNVIQMRR46 pKa = 3.81
MM1 pKa = 7.63ALRR4 pKa = 11.84RR5 pKa = 11.84HH6 pKa = 4.7THH8 pKa = 7.05ALGLWHH14 pKa = 6.39ATVRR18 pKa = 11.84ALKK21 pKa = 10.4IGFQNKK27 pKa = 8.82NMLFAKK33 pKa = 9.17PRR35 pKa = 11.84LMFRR39 pKa = 11.84QNVIQMRR46 pKa = 3.81
Molecular weight: 5.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
683258 |
13 |
2288 |
299.3 |
33.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.953 ± 0.069 | 0.969 ± 0.02 |
5.18 ± 0.045 | 5.628 ± 0.054 |
4.159 ± 0.039 | 6.693 ± 0.056 |
2.676 ± 0.038 | 6.083 ± 0.039 |
5.501 ± 0.048 | 10.151 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.572 ± 0.028 | 4.921 ± 0.055 |
4.103 ± 0.038 | 5.371 ± 0.052 |
4.566 ± 0.042 | 5.462 ± 0.037 |
5.105 ± 0.046 | 6.661 ± 0.055 |
1.338 ± 0.022 | 2.907 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
