
Ralstonia phage RSY1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Peduovirinae; Aresaunavirus; Ralstonia virus RSY1
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
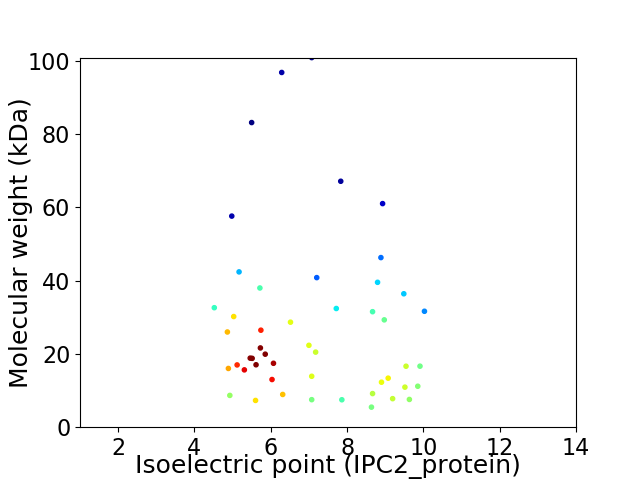
Virtual 2D-PAGE plot for 49 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
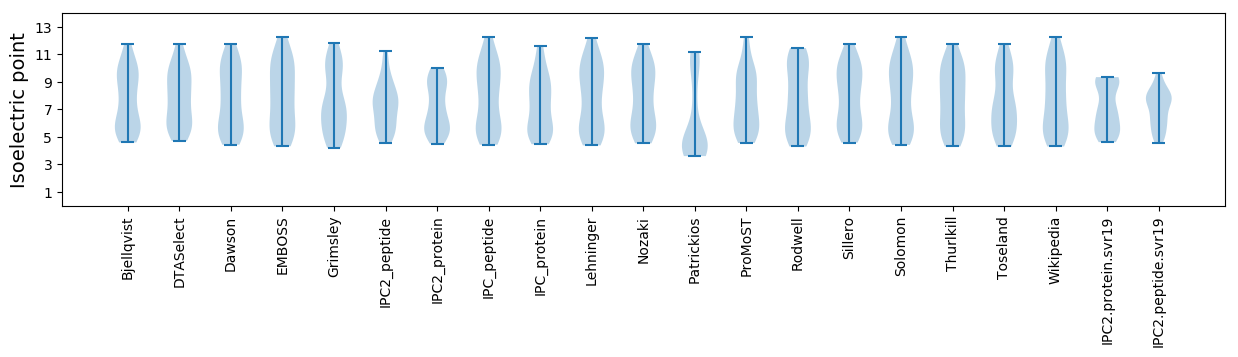
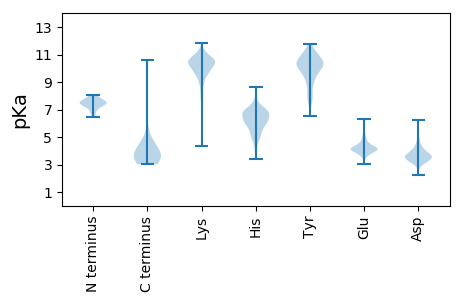
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A077K9Y4|A0A077K9Y4_9CAUD Uncharacterized protein OS=Ralstonia phage RSY1 OX=1530085 PE=3 SV=1
MM1 pKa = 7.26ATIDD5 pKa = 4.53LSQLPAPAVVEE16 pKa = 4.17TLDD19 pKa = 3.58YY20 pKa = 10.97EE21 pKa = 5.09AILAEE26 pKa = 4.14RR27 pKa = 11.84KK28 pKa = 10.13DD29 pKa = 4.24YY30 pKa = 10.68FVSLYY35 pKa = 10.33PADD38 pKa = 3.38QRR40 pKa = 11.84DD41 pKa = 3.64AVRR44 pKa = 11.84ATLEE48 pKa = 4.38LEE50 pKa = 4.22SEE52 pKa = 4.97PITKK56 pKa = 10.36LLQEE60 pKa = 3.83NAYY63 pKa = 10.37RR64 pKa = 11.84EE65 pKa = 4.4LVWRR69 pKa = 11.84QRR71 pKa = 11.84VNDD74 pKa = 3.86AARR77 pKa = 11.84GVMLAFAEE85 pKa = 4.78GEE87 pKa = 4.05DD88 pKa = 4.37LEE90 pKa = 5.07QIAANFNVQRR100 pKa = 11.84LTITPADD107 pKa = 4.13DD108 pKa = 3.49TTVPPTPAVMEE119 pKa = 4.7GDD121 pKa = 3.37DD122 pKa = 4.08SLRR125 pKa = 11.84EE126 pKa = 3.81RR127 pKa = 11.84AQEE130 pKa = 3.88AFEE133 pKa = 4.36GLSVAGPTKK142 pKa = 10.29AYY144 pKa = 9.83EE145 pKa = 4.0QFARR149 pKa = 11.84SADD152 pKa = 3.31GRR154 pKa = 11.84VADD157 pKa = 4.86ARR159 pKa = 11.84AISPAGAEE167 pKa = 4.17VVVSVLSHH175 pKa = 6.7LNDD178 pKa = 3.36GTADD182 pKa = 3.56EE183 pKa = 4.84SLLATVRR190 pKa = 11.84TALSDD195 pKa = 3.81DD196 pKa = 3.66DD197 pKa = 4.2TRR199 pKa = 11.84PLGDD203 pKa = 3.4RR204 pKa = 11.84LTVQSATIVPYY215 pKa = 10.48RR216 pKa = 11.84IRR218 pKa = 11.84ATLYY222 pKa = 10.2LASGPAAEE230 pKa = 5.8PILDD234 pKa = 3.94AAGKK238 pKa = 10.13RR239 pKa = 11.84ADD241 pKa = 3.48TYY243 pKa = 8.62RR244 pKa = 11.84TTRR247 pKa = 11.84RR248 pKa = 11.84RR249 pKa = 11.84IGRR252 pKa = 11.84DD253 pKa = 2.83INRR256 pKa = 11.84SAITAALHH264 pKa = 5.37VEE266 pKa = 4.34GVEE269 pKa = 3.92KK270 pKa = 10.8LVLHH274 pKa = 6.68EE275 pKa = 4.3PAEE278 pKa = 4.85DD279 pKa = 3.24IALDD283 pKa = 3.93LTQAGYY289 pKa = 8.4CTGVDD294 pKa = 3.11IVNGGTSEE302 pKa = 3.94
MM1 pKa = 7.26ATIDD5 pKa = 4.53LSQLPAPAVVEE16 pKa = 4.17TLDD19 pKa = 3.58YY20 pKa = 10.97EE21 pKa = 5.09AILAEE26 pKa = 4.14RR27 pKa = 11.84KK28 pKa = 10.13DD29 pKa = 4.24YY30 pKa = 10.68FVSLYY35 pKa = 10.33PADD38 pKa = 3.38QRR40 pKa = 11.84DD41 pKa = 3.64AVRR44 pKa = 11.84ATLEE48 pKa = 4.38LEE50 pKa = 4.22SEE52 pKa = 4.97PITKK56 pKa = 10.36LLQEE60 pKa = 3.83NAYY63 pKa = 10.37RR64 pKa = 11.84EE65 pKa = 4.4LVWRR69 pKa = 11.84QRR71 pKa = 11.84VNDD74 pKa = 3.86AARR77 pKa = 11.84GVMLAFAEE85 pKa = 4.78GEE87 pKa = 4.05DD88 pKa = 4.37LEE90 pKa = 5.07QIAANFNVQRR100 pKa = 11.84LTITPADD107 pKa = 4.13DD108 pKa = 3.49TTVPPTPAVMEE119 pKa = 4.7GDD121 pKa = 3.37DD122 pKa = 4.08SLRR125 pKa = 11.84EE126 pKa = 3.81RR127 pKa = 11.84AQEE130 pKa = 3.88AFEE133 pKa = 4.36GLSVAGPTKK142 pKa = 10.29AYY144 pKa = 9.83EE145 pKa = 4.0QFARR149 pKa = 11.84SADD152 pKa = 3.31GRR154 pKa = 11.84VADD157 pKa = 4.86ARR159 pKa = 11.84AISPAGAEE167 pKa = 4.17VVVSVLSHH175 pKa = 6.7LNDD178 pKa = 3.36GTADD182 pKa = 3.56EE183 pKa = 4.84SLLATVRR190 pKa = 11.84TALSDD195 pKa = 3.81DD196 pKa = 3.66DD197 pKa = 4.2TRR199 pKa = 11.84PLGDD203 pKa = 3.4RR204 pKa = 11.84LTVQSATIVPYY215 pKa = 10.48RR216 pKa = 11.84IRR218 pKa = 11.84ATLYY222 pKa = 10.2LASGPAAEE230 pKa = 5.8PILDD234 pKa = 3.94AAGKK238 pKa = 10.13RR239 pKa = 11.84ADD241 pKa = 3.48TYY243 pKa = 8.62RR244 pKa = 11.84TTRR247 pKa = 11.84RR248 pKa = 11.84RR249 pKa = 11.84IGRR252 pKa = 11.84DD253 pKa = 2.83INRR256 pKa = 11.84SAITAALHH264 pKa = 5.37VEE266 pKa = 4.34GVEE269 pKa = 3.92KK270 pKa = 10.8LVLHH274 pKa = 6.68EE275 pKa = 4.3PAEE278 pKa = 4.85DD279 pKa = 3.24IALDD283 pKa = 3.93LTQAGYY289 pKa = 8.4CTGVDD294 pKa = 3.11IVNGGTSEE302 pKa = 3.94
Molecular weight: 32.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A077K9Q8|A0A077K9Q8_9CAUD Uncharacterized protein OS=Ralstonia phage RSY1 OX=1530085 PE=3 SV=1
MM1 pKa = 7.55RR2 pKa = 11.84RR3 pKa = 11.84CPCLYY8 pKa = 9.37PVPGTRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84LSLAAQAGEE25 pKa = 4.32APTGAAYY32 pKa = 8.86WCICCEE38 pKa = 4.95LLTKK42 pKa = 9.19WAPRR46 pKa = 11.84GSALQIGTKK55 pKa = 10.07VPLRR59 pKa = 11.84GFSRR63 pKa = 11.84MVKK66 pKa = 10.07LRR68 pKa = 11.84ADD70 pKa = 3.5NKK72 pKa = 10.22EE73 pKa = 3.84GEE75 pKa = 4.16FPIIRR80 pKa = 11.84KK81 pKa = 8.67VFNLSPGLIFRR92 pKa = 11.84LLNIAHH98 pKa = 7.26
MM1 pKa = 7.55RR2 pKa = 11.84RR3 pKa = 11.84CPCLYY8 pKa = 9.37PVPGTRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84LSLAAQAGEE25 pKa = 4.32APTGAAYY32 pKa = 8.86WCICCEE38 pKa = 4.95LLTKK42 pKa = 9.19WAPRR46 pKa = 11.84GSALQIGTKK55 pKa = 10.07VPLRR59 pKa = 11.84GFSRR63 pKa = 11.84MVKK66 pKa = 10.07LRR68 pKa = 11.84ADD70 pKa = 3.5NKK72 pKa = 10.22EE73 pKa = 3.84GEE75 pKa = 4.16FPIIRR80 pKa = 11.84KK81 pKa = 8.67VFNLSPGLIFRR92 pKa = 11.84LLNIAHH98 pKa = 7.26
Molecular weight: 10.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12425 |
55 |
901 |
253.6 |
27.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.797 ± 0.521 | 0.934 ± 0.129 |
5.795 ± 0.197 | 5.416 ± 0.231 |
3.219 ± 0.193 | 7.831 ± 0.39 |
2.254 ± 0.26 | 3.903 ± 0.169 |
3.855 ± 0.258 | 8.869 ± 0.247 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.157 ± 0.136 | 3.292 ± 0.199 |
4.909 ± 0.219 | 4.209 ± 0.216 |
7.863 ± 0.437 | 5.078 ± 0.219 |
5.755 ± 0.376 | 7.509 ± 0.252 |
1.65 ± 0.13 | 2.704 ± 0.19 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
