
Southern bean mosaic virus (isolate Bean/United States/Arkansas) (SBMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus; Southern bean mosaic virus
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
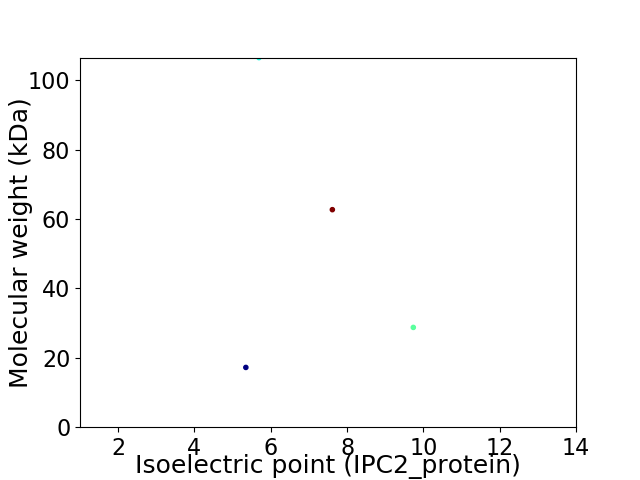
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
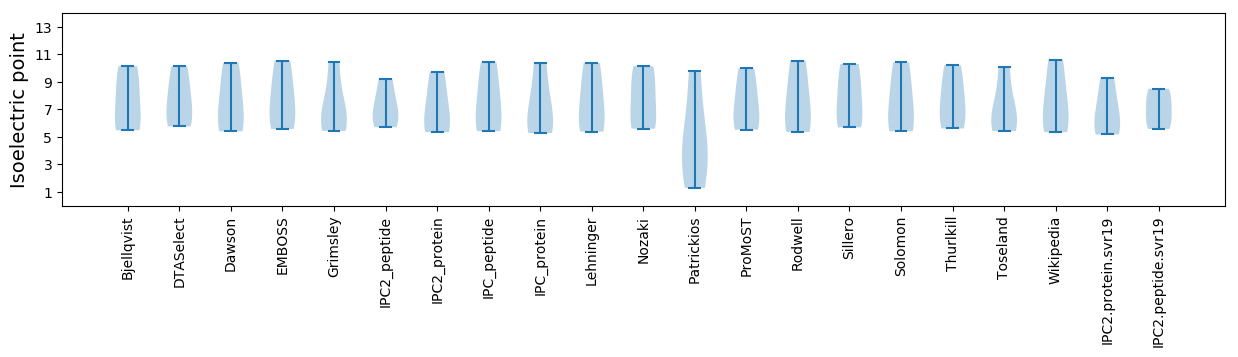
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|O73571|MVP_SBMVA Movement protein P1 OS=Southern bean mosaic virus (isolate Bean/United States/Arkansas) OX=652938 GN=ORF1 PE=3 SV=1
MM1 pKa = 7.91SYY3 pKa = 10.9RR4 pKa = 11.84FLTVRR9 pKa = 11.84AFGFTGFHH17 pKa = 6.97CDD19 pKa = 2.89ATRR22 pKa = 11.84LLSEE26 pKa = 4.57TEE28 pKa = 4.32VIDD31 pKa = 4.1VPSSLDD37 pKa = 3.5FVGEE41 pKa = 4.15TEE43 pKa = 5.27LRR45 pKa = 11.84LEE47 pKa = 4.47TAWPQCEE54 pKa = 4.19EE55 pKa = 3.82NCYY58 pKa = 10.2TILPRR63 pKa = 11.84FNVQVDD69 pKa = 3.94FEE71 pKa = 4.84YY72 pKa = 11.17YY73 pKa = 8.29PVRR76 pKa = 11.84VEE78 pKa = 3.87IVCRR82 pKa = 11.84VCAASLSVIFSKK94 pKa = 10.5WDD96 pKa = 4.2FYY98 pKa = 11.06CSRR101 pKa = 11.84RR102 pKa = 11.84GHH104 pKa = 6.15FVPVDD109 pKa = 3.46QNGDD113 pKa = 3.49LFRR116 pKa = 11.84IGTLQEE122 pKa = 3.75TGEE125 pKa = 4.33KK126 pKa = 10.56YY127 pKa = 10.47FYY129 pKa = 10.88FCDD132 pKa = 3.9KK133 pKa = 10.67SICRR137 pKa = 11.84QCIIQAAHH145 pKa = 5.75HH146 pKa = 6.36HH147 pKa = 5.58SS148 pKa = 3.48
MM1 pKa = 7.91SYY3 pKa = 10.9RR4 pKa = 11.84FLTVRR9 pKa = 11.84AFGFTGFHH17 pKa = 6.97CDD19 pKa = 2.89ATRR22 pKa = 11.84LLSEE26 pKa = 4.57TEE28 pKa = 4.32VIDD31 pKa = 4.1VPSSLDD37 pKa = 3.5FVGEE41 pKa = 4.15TEE43 pKa = 5.27LRR45 pKa = 11.84LEE47 pKa = 4.47TAWPQCEE54 pKa = 4.19EE55 pKa = 3.82NCYY58 pKa = 10.2TILPRR63 pKa = 11.84FNVQVDD69 pKa = 3.94FEE71 pKa = 4.84YY72 pKa = 11.17YY73 pKa = 8.29PVRR76 pKa = 11.84VEE78 pKa = 3.87IVCRR82 pKa = 11.84VCAASLSVIFSKK94 pKa = 10.5WDD96 pKa = 4.2FYY98 pKa = 11.06CSRR101 pKa = 11.84RR102 pKa = 11.84GHH104 pKa = 6.15FVPVDD109 pKa = 3.46QNGDD113 pKa = 3.49LFRR116 pKa = 11.84IGTLQEE122 pKa = 3.75TGEE125 pKa = 4.33KK126 pKa = 10.56YY127 pKa = 10.47FYY129 pKa = 10.88FCDD132 pKa = 3.9KK133 pKa = 10.67SICRR137 pKa = 11.84QCIIQAAHH145 pKa = 5.75HH146 pKa = 6.36HH147 pKa = 5.58SS148 pKa = 3.48
Molecular weight: 17.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|O73564|P2A_SBMVA Polyprotein P2A OS=Southern bean mosaic virus (isolate Bean/United States/Arkansas) OX=652938 GN=ORF2A PE=4 SV=2
MM1 pKa = 7.87AKK3 pKa = 10.33RR4 pKa = 11.84LTKK7 pKa = 10.27QQLTKK12 pKa = 10.82AIANTLEE19 pKa = 4.14APATQSRR26 pKa = 11.84RR27 pKa = 11.84PRR29 pKa = 11.84NRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SAARR39 pKa = 11.84QPQSTQAGVSMAPIAQGTMVRR60 pKa = 11.84LRR62 pKa = 11.84EE63 pKa = 4.0PSLRR67 pKa = 11.84TAGGVTVLTHH77 pKa = 6.43SEE79 pKa = 4.11LSTEE83 pKa = 4.12LSVTNAIVITSEE95 pKa = 4.02LVMPYY100 pKa = 10.63TMGTWLRR107 pKa = 11.84GVAANWSKK115 pKa = 11.51YY116 pKa = 9.42SLLSVRR122 pKa = 11.84YY123 pKa = 7.52TYY125 pKa = 11.0LPSCPSTTSGSIHH138 pKa = 6.61MGFQYY143 pKa = 11.56DD144 pKa = 3.61MADD147 pKa = 3.6TLPVSVNQLSNLRR160 pKa = 11.84GYY162 pKa = 11.09VSGQVWSGSSGLCYY176 pKa = 10.31INGTRR181 pKa = 11.84CLDD184 pKa = 3.41TANAITTTLDD194 pKa = 2.86VGQLGKK200 pKa = 10.22KK201 pKa = 8.7WYY203 pKa = 9.03PFKK206 pKa = 10.71TSTDD210 pKa = 3.44FTTAVGVNVNIATPLVPARR229 pKa = 11.84LIIAMLDD236 pKa = 3.78GSSSTAVSTGRR247 pKa = 11.84LYY249 pKa = 11.22VSYY252 pKa = 9.44TIQLIEE258 pKa = 4.11PTALALNNN266 pKa = 3.79
MM1 pKa = 7.87AKK3 pKa = 10.33RR4 pKa = 11.84LTKK7 pKa = 10.27QQLTKK12 pKa = 10.82AIANTLEE19 pKa = 4.14APATQSRR26 pKa = 11.84RR27 pKa = 11.84PRR29 pKa = 11.84NRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SAARR39 pKa = 11.84QPQSTQAGVSMAPIAQGTMVRR60 pKa = 11.84LRR62 pKa = 11.84EE63 pKa = 4.0PSLRR67 pKa = 11.84TAGGVTVLTHH77 pKa = 6.43SEE79 pKa = 4.11LSTEE83 pKa = 4.12LSVTNAIVITSEE95 pKa = 4.02LVMPYY100 pKa = 10.63TMGTWLRR107 pKa = 11.84GVAANWSKK115 pKa = 11.51YY116 pKa = 9.42SLLSVRR122 pKa = 11.84YY123 pKa = 7.52TYY125 pKa = 11.0LPSCPSTTSGSIHH138 pKa = 6.61MGFQYY143 pKa = 11.56DD144 pKa = 3.61MADD147 pKa = 3.6TLPVSVNQLSNLRR160 pKa = 11.84GYY162 pKa = 11.09VSGQVWSGSSGLCYY176 pKa = 10.31INGTRR181 pKa = 11.84CLDD184 pKa = 3.41TANAITTTLDD194 pKa = 2.86VGQLGKK200 pKa = 10.22KK201 pKa = 8.7WYY203 pKa = 9.03PFKK206 pKa = 10.71TSTDD210 pKa = 3.44FTTAVGVNVNIATPLVPARR229 pKa = 11.84LIIAMLDD236 pKa = 3.78GSSSTAVSTGRR247 pKa = 11.84LYY249 pKa = 11.22VSYY252 pKa = 9.44TIQLIEE258 pKa = 4.11PTALALNNN266 pKa = 3.79
Molecular weight: 28.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1951 |
148 |
962 |
487.8 |
53.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.176 ± 0.516 | 2.665 ± 0.503 |
4.562 ± 0.64 | 5.843 ± 0.931 |
3.69 ± 0.754 | 7.227 ± 0.437 |
1.896 ± 0.361 | 4.1 ± 0.266 |
5.126 ± 0.948 | 9.124 ± 0.328 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.794 ± 0.301 | 2.87 ± 0.403 |
6.458 ± 0.913 | 3.537 ± 0.362 |
5.382 ± 0.684 | 9.995 ± 0.579 |
5.792 ± 1.717 | 7.381 ± 0.206 |
2.101 ± 0.268 | 3.28 ± 0.269 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
