
Heterobasidion irregulare (strain TC 32-1)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Russulales; Bondarzewiaceae; Heterobasidion; Heterobasidion annosum species complex; Heterobasidion irregulare
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
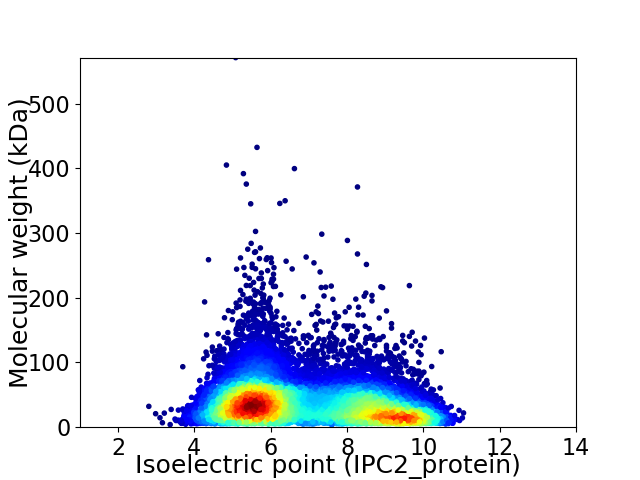
Virtual 2D-PAGE plot for 13159 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W4KID9|W4KID9_HETIT Phosphoglycerate kinase OS=Heterobasidion irregulare (strain TC 32-1) OX=747525 GN=HETIRDRAFT_407577 PE=3 SV=1
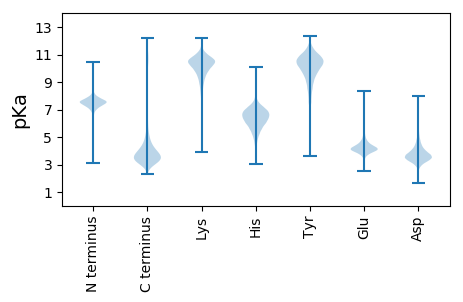
MM1 pKa = 7.26FPSPSWLDD9 pKa = 3.84DD10 pKa = 4.19DD11 pKa = 5.96DD12 pKa = 7.29ASTLDD17 pKa = 4.01NNASTLDD24 pKa = 3.96DD25 pKa = 4.01NASLLDD31 pKa = 3.99NNDD34 pKa = 3.61AGHH37 pKa = 6.69FSHH40 pKa = 7.2SGTCSLAPSASDD52 pKa = 5.41LDD54 pKa = 4.45LDD56 pKa = 4.01PCGLLGTPEE65 pKa = 4.17PTLSLSDD72 pKa = 3.98DD73 pKa = 3.88EE74 pKa = 5.15EE75 pKa = 5.33PIPEE79 pKa = 4.61LKK81 pKa = 10.79NNPQYY86 pKa = 9.51QTSEE90 pKa = 3.92EE91 pKa = 4.1LFEE94 pKa = 4.88CFLDD98 pKa = 4.11SGSEE102 pKa = 3.95HH103 pKa = 7.55LDD105 pKa = 3.69DD106 pKa = 5.54NNSNPPAFDD115 pKa = 4.34KK116 pKa = 11.34DD117 pKa = 3.58PAICNAYY124 pKa = 7.44ITAYY128 pKa = 10.9LLTACHH134 pKa = 6.9RR135 pKa = 11.84STQEE139 pKa = 3.86AIKK142 pKa = 10.37AYY144 pKa = 10.62LDD146 pKa = 4.06AEE148 pKa = 4.32HH149 pKa = 6.97EE150 pKa = 4.5MLLFMQRR157 pKa = 11.84RR158 pKa = 11.84TGLEE162 pKa = 3.93MPGLDD167 pKa = 3.2TMAWTLLTLEE177 pKa = 4.44
MM1 pKa = 7.26FPSPSWLDD9 pKa = 3.84DD10 pKa = 4.19DD11 pKa = 5.96DD12 pKa = 7.29ASTLDD17 pKa = 4.01NNASTLDD24 pKa = 3.96DD25 pKa = 4.01NASLLDD31 pKa = 3.99NNDD34 pKa = 3.61AGHH37 pKa = 6.69FSHH40 pKa = 7.2SGTCSLAPSASDD52 pKa = 5.41LDD54 pKa = 4.45LDD56 pKa = 4.01PCGLLGTPEE65 pKa = 4.17PTLSLSDD72 pKa = 3.98DD73 pKa = 3.88EE74 pKa = 5.15EE75 pKa = 5.33PIPEE79 pKa = 4.61LKK81 pKa = 10.79NNPQYY86 pKa = 9.51QTSEE90 pKa = 3.92EE91 pKa = 4.1LFEE94 pKa = 4.88CFLDD98 pKa = 4.11SGSEE102 pKa = 3.95HH103 pKa = 7.55LDD105 pKa = 3.69DD106 pKa = 5.54NNSNPPAFDD115 pKa = 4.34KK116 pKa = 11.34DD117 pKa = 3.58PAICNAYY124 pKa = 7.44ITAYY128 pKa = 10.9LLTACHH134 pKa = 6.9RR135 pKa = 11.84STQEE139 pKa = 3.86AIKK142 pKa = 10.37AYY144 pKa = 10.62LDD146 pKa = 4.06AEE148 pKa = 4.32HH149 pKa = 6.97EE150 pKa = 4.5MLLFMQRR157 pKa = 11.84RR158 pKa = 11.84TGLEE162 pKa = 3.93MPGLDD167 pKa = 3.2TMAWTLLTLEE177 pKa = 4.44
Molecular weight: 19.42 kDa
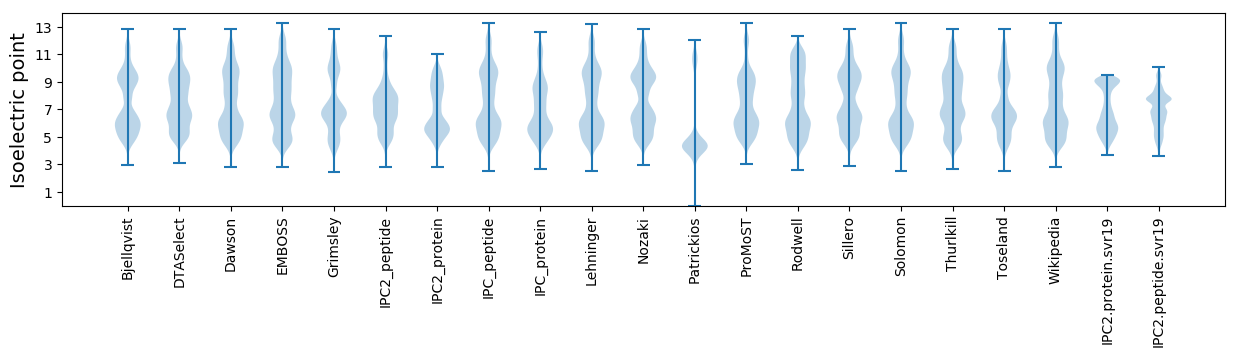
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W4JPK7|W4JPK7_HETIT Uncharacterized protein OS=Heterobasidion irregulare (strain TC 32-1) OX=747525 GN=HETIRDRAFT_442718 PE=4 SV=1
MM1 pKa = 7.53GSSCTARR8 pKa = 11.84CPPRR12 pKa = 11.84CPPRR16 pKa = 11.84RR17 pKa = 11.84LDD19 pKa = 3.36RR20 pKa = 11.84PSRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84VRR28 pKa = 11.84EE29 pKa = 3.56RR30 pKa = 11.84RR31 pKa = 11.84HH32 pKa = 5.61RR33 pKa = 11.84APLPRR38 pKa = 11.84SRR40 pKa = 11.84PSRR43 pKa = 11.84RR44 pKa = 11.84PPPRR48 pKa = 11.84AHH50 pKa = 6.54SPNLPRR56 pKa = 11.84PSPRR60 pKa = 11.84PRR62 pKa = 11.84TRR64 pKa = 11.84SSPRR68 pKa = 11.84PRR70 pKa = 11.84APRR73 pKa = 11.84APRR76 pKa = 11.84THH78 pKa = 6.42TRR80 pKa = 11.84PAIPTPSPRR89 pKa = 11.84TRR91 pKa = 11.84TRR93 pKa = 11.84THH95 pKa = 5.72TRR97 pKa = 11.84THH99 pKa = 4.95TRR101 pKa = 11.84RR102 pKa = 11.84SRR104 pKa = 11.84ARR106 pKa = 11.84RR107 pKa = 11.84SASRR111 pKa = 11.84GRR113 pKa = 11.84SRR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84GMGRR121 pKa = 11.84PTGRR125 pKa = 11.84GTGRR129 pKa = 11.84ATTHH133 pKa = 6.11RR134 pKa = 11.84AIMPRR139 pKa = 11.84RR140 pKa = 11.84ALARR144 pKa = 11.84RR145 pKa = 11.84ALTRR149 pKa = 11.84RR150 pKa = 11.84GTARR154 pKa = 11.84RR155 pKa = 11.84GTARR159 pKa = 11.84RR160 pKa = 11.84GTARR164 pKa = 11.84RR165 pKa = 11.84GIMGRR170 pKa = 11.84DD171 pKa = 2.9TGRR174 pKa = 11.84RR175 pKa = 11.84IRR177 pKa = 11.84SRR179 pKa = 11.84RR180 pKa = 11.84CCRR183 pKa = 11.84RR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84RR187 pKa = 11.84RR188 pKa = 11.84GG189 pKa = 3.04
MM1 pKa = 7.53GSSCTARR8 pKa = 11.84CPPRR12 pKa = 11.84CPPRR16 pKa = 11.84RR17 pKa = 11.84LDD19 pKa = 3.36RR20 pKa = 11.84PSRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84VRR28 pKa = 11.84EE29 pKa = 3.56RR30 pKa = 11.84RR31 pKa = 11.84HH32 pKa = 5.61RR33 pKa = 11.84APLPRR38 pKa = 11.84SRR40 pKa = 11.84PSRR43 pKa = 11.84RR44 pKa = 11.84PPPRR48 pKa = 11.84AHH50 pKa = 6.54SPNLPRR56 pKa = 11.84PSPRR60 pKa = 11.84PRR62 pKa = 11.84TRR64 pKa = 11.84SSPRR68 pKa = 11.84PRR70 pKa = 11.84APRR73 pKa = 11.84APRR76 pKa = 11.84THH78 pKa = 6.42TRR80 pKa = 11.84PAIPTPSPRR89 pKa = 11.84TRR91 pKa = 11.84TRR93 pKa = 11.84THH95 pKa = 5.72TRR97 pKa = 11.84THH99 pKa = 4.95TRR101 pKa = 11.84RR102 pKa = 11.84SRR104 pKa = 11.84ARR106 pKa = 11.84RR107 pKa = 11.84SASRR111 pKa = 11.84GRR113 pKa = 11.84SRR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84GMGRR121 pKa = 11.84PTGRR125 pKa = 11.84GTGRR129 pKa = 11.84ATTHH133 pKa = 6.11RR134 pKa = 11.84AIMPRR139 pKa = 11.84RR140 pKa = 11.84ALARR144 pKa = 11.84RR145 pKa = 11.84ALTRR149 pKa = 11.84RR150 pKa = 11.84GTARR154 pKa = 11.84RR155 pKa = 11.84GTARR159 pKa = 11.84RR160 pKa = 11.84GTARR164 pKa = 11.84RR165 pKa = 11.84GIMGRR170 pKa = 11.84DD171 pKa = 2.9TGRR174 pKa = 11.84RR175 pKa = 11.84IRR177 pKa = 11.84SRR179 pKa = 11.84RR180 pKa = 11.84CCRR183 pKa = 11.84RR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84RR187 pKa = 11.84RR188 pKa = 11.84GG189 pKa = 3.04
Molecular weight: 22.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5008163 |
30 |
5124 |
380.6 |
41.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.193 ± 0.025 | 1.271 ± 0.01 |
5.55 ± 0.017 | 5.61 ± 0.021 |
3.654 ± 0.014 | 6.602 ± 0.021 |
2.738 ± 0.012 | 4.712 ± 0.02 |
4.129 ± 0.019 | 9.247 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.079 ± 0.008 | 3.16 ± 0.014 |
6.744 ± 0.035 | 3.614 ± 0.016 |
6.766 ± 0.024 | 8.666 ± 0.028 |
5.907 ± 0.015 | 6.37 ± 0.017 |
1.424 ± 0.008 | 2.564 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
