
Dipodomys ordii (Ord s kangaroo rat)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria; Boreoeutheria; Euarchontoglires; Glires; Rodentia
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
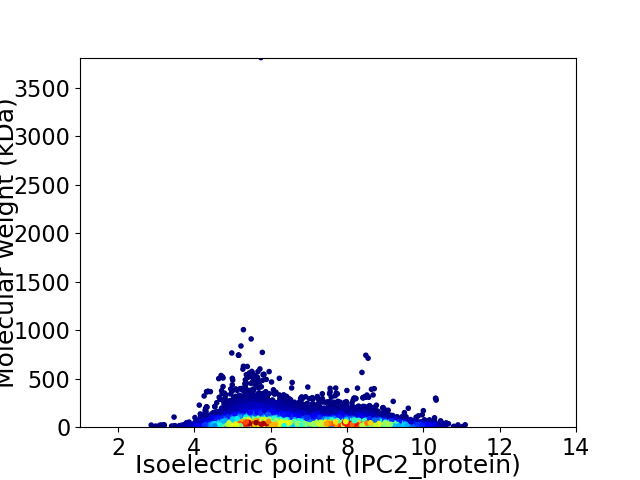
Virtual 2D-PAGE plot for 26567 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
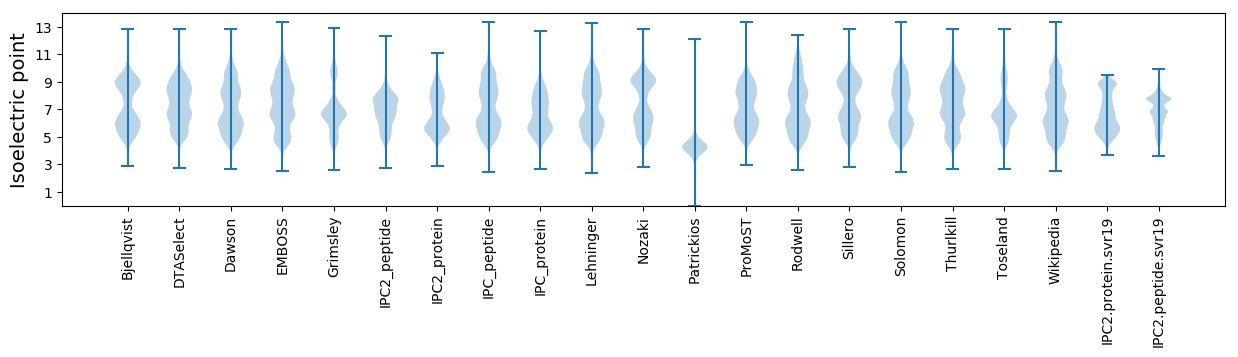
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S3GI92|A0A1S3GI92_DIPOR Protein-arginine deiminase OS=Dipodomys ordii OX=10020 GN=Padi1 PE=3 SV=1
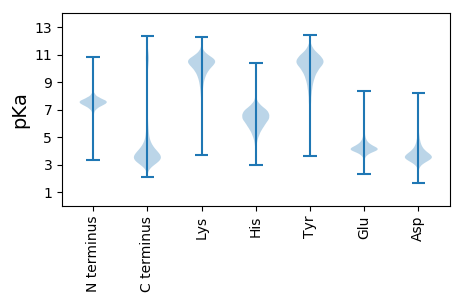
MM1 pKa = 7.58TEE3 pKa = 3.84TSVTEE8 pKa = 3.96TSVTEE13 pKa = 4.1TSVTDD18 pKa = 3.59PSMTEE23 pKa = 3.77TSVTGQCDD31 pKa = 3.29RR32 pKa = 11.84DD33 pKa = 3.5QCDD36 pKa = 3.32RR37 pKa = 11.84DD38 pKa = 3.47QCDD41 pKa = 3.29RR42 pKa = 11.84SHH44 pKa = 5.81VTDD47 pKa = 4.68PSVTDD52 pKa = 3.65PSVTEE57 pKa = 4.03TSVTDD62 pKa = 3.68PSVTEE67 pKa = 3.89TSVTEE72 pKa = 4.0TSVTEE77 pKa = 4.0TSVTEE82 pKa = 4.0TSVTEE87 pKa = 4.1TSVTDD92 pKa = 3.68PSVTEE97 pKa = 4.03TSVTDD102 pKa = 3.68PSVTEE107 pKa = 3.89TSVTEE112 pKa = 4.0TSVTEE117 pKa = 4.1TSVTDD122 pKa = 3.85PSVTDD127 pKa = 3.98PSVTDD132 pKa = 3.98PSVTDD137 pKa = 3.98PSVTDD142 pKa = 3.72PSVTDD147 pKa = 3.16QCDD150 pKa = 3.18RR151 pKa = 11.84PQCDD155 pKa = 2.55RR156 pKa = 11.84GQCHH160 pKa = 6.84RR161 pKa = 11.84GQCHH165 pKa = 6.57RR166 pKa = 11.84GQCHH170 pKa = 6.27RR171 pKa = 11.84PQCDD175 pKa = 3.4GDD177 pKa = 3.75PAAPTFIDD185 pKa = 3.76TLDD188 pKa = 3.72CALAA192 pKa = 4.47
MM1 pKa = 7.58TEE3 pKa = 3.84TSVTEE8 pKa = 3.96TSVTEE13 pKa = 4.1TSVTDD18 pKa = 3.59PSMTEE23 pKa = 3.77TSVTGQCDD31 pKa = 3.29RR32 pKa = 11.84DD33 pKa = 3.5QCDD36 pKa = 3.32RR37 pKa = 11.84DD38 pKa = 3.47QCDD41 pKa = 3.29RR42 pKa = 11.84SHH44 pKa = 5.81VTDD47 pKa = 4.68PSVTDD52 pKa = 3.65PSVTEE57 pKa = 4.03TSVTDD62 pKa = 3.68PSVTEE67 pKa = 3.89TSVTEE72 pKa = 4.0TSVTEE77 pKa = 4.0TSVTEE82 pKa = 4.0TSVTEE87 pKa = 4.1TSVTDD92 pKa = 3.68PSVTEE97 pKa = 4.03TSVTDD102 pKa = 3.68PSVTEE107 pKa = 3.89TSVTEE112 pKa = 4.0TSVTEE117 pKa = 4.1TSVTDD122 pKa = 3.85PSVTDD127 pKa = 3.98PSVTDD132 pKa = 3.98PSVTDD137 pKa = 3.98PSVTDD142 pKa = 3.72PSVTDD147 pKa = 3.16QCDD150 pKa = 3.18RR151 pKa = 11.84PQCDD155 pKa = 2.55RR156 pKa = 11.84GQCHH160 pKa = 6.84RR161 pKa = 11.84GQCHH165 pKa = 6.57RR166 pKa = 11.84GQCHH170 pKa = 6.27RR171 pKa = 11.84PQCDD175 pKa = 3.4GDD177 pKa = 3.75PAAPTFIDD185 pKa = 3.76TLDD188 pKa = 3.72CALAA192 pKa = 4.47
Molecular weight: 20.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S3FZU8|A0A1S3FZU8_DIPOR stimulated by retinoic acid gene 6 protein homolog OS=Dipodomys ordii OX=10020 GN=LOC105993383 PE=4 SV=1
MM1 pKa = 7.11SQNPQRR7 pKa = 11.84RR8 pKa = 11.84APSPLPWKK16 pKa = 8.91RR17 pKa = 11.84TSIKK21 pKa = 9.75PPRR24 pKa = 11.84PSTHH28 pKa = 5.92TPRR31 pKa = 11.84TPHH34 pKa = 6.38PAPRR38 pKa = 11.84TPHH41 pKa = 7.09PDD43 pKa = 3.06SAPRR47 pKa = 11.84PLPHH51 pKa = 7.17PSASRR56 pKa = 11.84CRR58 pKa = 11.84SGRR61 pKa = 11.84PGRR64 pKa = 11.84ARR66 pKa = 11.84TRR68 pKa = 11.84QALRR72 pKa = 11.84RR73 pKa = 11.84PPAAPQFLVEE83 pKa = 4.11ATVRR87 pKa = 11.84GPAKK91 pKa = 10.06PPLPRR96 pKa = 11.84TPAVAHH102 pKa = 6.0AHH104 pKa = 5.81RR105 pKa = 11.84PGHH108 pKa = 5.96RR109 pKa = 11.84MTRR112 pKa = 11.84TTPGHH117 pKa = 5.05TRR119 pKa = 11.84RR120 pKa = 11.84PGPGRR125 pKa = 11.84CRR127 pKa = 11.84ASSQTLTRR135 pKa = 11.84RR136 pKa = 11.84SRR138 pKa = 11.84LPWPPPRR145 pKa = 11.84PHH147 pKa = 7.11ARR149 pKa = 11.84RR150 pKa = 11.84LLPRR154 pKa = 11.84GGPGRR159 pKa = 11.84CSFPARR165 pKa = 11.84PPSGARR171 pKa = 11.84RR172 pKa = 11.84AKK174 pKa = 9.67PRR176 pKa = 11.84SGVSRR181 pKa = 11.84RR182 pKa = 11.84RR183 pKa = 11.84RR184 pKa = 11.84RR185 pKa = 11.84HH186 pKa = 4.35RR187 pKa = 11.84RR188 pKa = 11.84RR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84PLHH194 pKa = 5.45FTSLPPPSMRR204 pKa = 11.84PSHH207 pKa = 6.85RR208 pKa = 11.84GPPRR212 pKa = 11.84ISWIRR217 pKa = 11.84SRR219 pKa = 4.43
MM1 pKa = 7.11SQNPQRR7 pKa = 11.84RR8 pKa = 11.84APSPLPWKK16 pKa = 8.91RR17 pKa = 11.84TSIKK21 pKa = 9.75PPRR24 pKa = 11.84PSTHH28 pKa = 5.92TPRR31 pKa = 11.84TPHH34 pKa = 6.38PAPRR38 pKa = 11.84TPHH41 pKa = 7.09PDD43 pKa = 3.06SAPRR47 pKa = 11.84PLPHH51 pKa = 7.17PSASRR56 pKa = 11.84CRR58 pKa = 11.84SGRR61 pKa = 11.84PGRR64 pKa = 11.84ARR66 pKa = 11.84TRR68 pKa = 11.84QALRR72 pKa = 11.84RR73 pKa = 11.84PPAAPQFLVEE83 pKa = 4.11ATVRR87 pKa = 11.84GPAKK91 pKa = 10.06PPLPRR96 pKa = 11.84TPAVAHH102 pKa = 6.0AHH104 pKa = 5.81RR105 pKa = 11.84PGHH108 pKa = 5.96RR109 pKa = 11.84MTRR112 pKa = 11.84TTPGHH117 pKa = 5.05TRR119 pKa = 11.84RR120 pKa = 11.84PGPGRR125 pKa = 11.84CRR127 pKa = 11.84ASSQTLTRR135 pKa = 11.84RR136 pKa = 11.84SRR138 pKa = 11.84LPWPPPRR145 pKa = 11.84PHH147 pKa = 7.11ARR149 pKa = 11.84RR150 pKa = 11.84LLPRR154 pKa = 11.84GGPGRR159 pKa = 11.84CSFPARR165 pKa = 11.84PPSGARR171 pKa = 11.84RR172 pKa = 11.84AKK174 pKa = 9.67PRR176 pKa = 11.84SGVSRR181 pKa = 11.84RR182 pKa = 11.84RR183 pKa = 11.84RR184 pKa = 11.84RR185 pKa = 11.84HH186 pKa = 4.35RR187 pKa = 11.84RR188 pKa = 11.84RR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84PLHH194 pKa = 5.45FTSLPPPSMRR204 pKa = 11.84PSHH207 pKa = 6.85RR208 pKa = 11.84GPPRR212 pKa = 11.84ISWIRR217 pKa = 11.84SRR219 pKa = 4.43
Molecular weight: 24.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
15473226 |
31 |
34334 |
582.4 |
64.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.921 ± 0.017 | 2.198 ± 0.013 |
4.802 ± 0.011 | 7.033 ± 0.021 |
3.675 ± 0.012 | 6.415 ± 0.019 |
2.608 ± 0.007 | 4.377 ± 0.015 |
5.681 ± 0.019 | 10.036 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.207 ± 0.006 | 3.615 ± 0.011 |
6.339 ± 0.025 | 4.817 ± 0.016 |
5.621 ± 0.014 | 8.436 ± 0.022 |
5.354 ± 0.014 | 6.023 ± 0.014 |
1.185 ± 0.005 | 2.65 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
