
Ogataea parapolymorpha (strain ATCC 26012 / BCRC 20466 / JCM 22074 / NRRL Y-7560 / DL-1) (Yeast) (Hansenula polymorpha)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Pichiaceae; Ogataea; Ogataea parapolymorpha
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
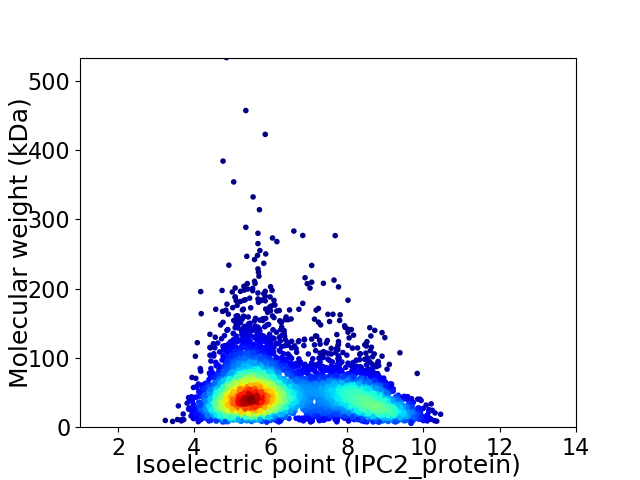
Virtual 2D-PAGE plot for 5338 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W1QK75|W1QK75_OGAPD Clathrin coat assembly protein OS=Ogataea parapolymorpha (strain ATCC 26012 / BCRC 20466 / JCM 22074 / NRRL Y-7560 / DL-1) OX=871575 GN=HPODL_05008 PE=4 SV=1
MM1 pKa = 7.39SLLRR5 pKa = 11.84NFIDD9 pKa = 3.61ALSPITAYY17 pKa = 10.48AEE19 pKa = 4.16EE20 pKa = 4.39PAEE23 pKa = 4.13EE24 pKa = 4.04PVEE27 pKa = 4.12EE28 pKa = 4.39VEE30 pKa = 4.51EE31 pKa = 4.22EE32 pKa = 4.06ASEE35 pKa = 4.27EE36 pKa = 4.06PQEE39 pKa = 4.16EE40 pKa = 4.38EE41 pKa = 4.88AGGDD45 pKa = 3.48DD46 pKa = 4.56AEE48 pKa = 4.56EE49 pKa = 4.32EE50 pKa = 4.39EE51 pKa = 5.49EE52 pKa = 5.36DD53 pKa = 5.66DD54 pKa = 5.82EE55 pKa = 7.17DD56 pKa = 6.64DD57 pKa = 4.69EE58 pKa = 7.21DD59 pKa = 5.7EE60 pKa = 5.25DD61 pKa = 6.4DD62 pKa = 4.16EE63 pKa = 6.26DD64 pKa = 4.84EE65 pKa = 4.41EE66 pKa = 6.43AGDD69 pKa = 4.48PMDD72 pKa = 5.44ALRR75 pKa = 11.84KK76 pKa = 8.97QCSEE80 pKa = 4.19TKK82 pKa = 9.12EE83 pKa = 4.07CAHH86 pKa = 6.11YY87 pKa = 8.95VHH89 pKa = 7.39HH90 pKa = 5.97YY91 pKa = 7.83QEE93 pKa = 4.48CVEE96 pKa = 4.21RR97 pKa = 11.84VTKK100 pKa = 10.43EE101 pKa = 3.85MEE103 pKa = 3.87EE104 pKa = 3.85PGYY107 pKa = 10.3DD108 pKa = 3.08EE109 pKa = 5.86KK110 pKa = 11.14EE111 pKa = 4.15YY112 pKa = 11.23KK113 pKa = 9.72EE114 pKa = 4.32DD115 pKa = 3.51CVEE118 pKa = 4.17EE119 pKa = 4.55FFHH122 pKa = 6.74LQHH125 pKa = 7.19CINDD129 pKa = 3.77CVAPKK134 pKa = 10.61LFYY137 pKa = 10.45KK138 pKa = 10.84LKK140 pKa = 10.57
MM1 pKa = 7.39SLLRR5 pKa = 11.84NFIDD9 pKa = 3.61ALSPITAYY17 pKa = 10.48AEE19 pKa = 4.16EE20 pKa = 4.39PAEE23 pKa = 4.13EE24 pKa = 4.04PVEE27 pKa = 4.12EE28 pKa = 4.39VEE30 pKa = 4.51EE31 pKa = 4.22EE32 pKa = 4.06ASEE35 pKa = 4.27EE36 pKa = 4.06PQEE39 pKa = 4.16EE40 pKa = 4.38EE41 pKa = 4.88AGGDD45 pKa = 3.48DD46 pKa = 4.56AEE48 pKa = 4.56EE49 pKa = 4.32EE50 pKa = 4.39EE51 pKa = 5.49EE52 pKa = 5.36DD53 pKa = 5.66DD54 pKa = 5.82EE55 pKa = 7.17DD56 pKa = 6.64DD57 pKa = 4.69EE58 pKa = 7.21DD59 pKa = 5.7EE60 pKa = 5.25DD61 pKa = 6.4DD62 pKa = 4.16EE63 pKa = 6.26DD64 pKa = 4.84EE65 pKa = 4.41EE66 pKa = 6.43AGDD69 pKa = 4.48PMDD72 pKa = 5.44ALRR75 pKa = 11.84KK76 pKa = 8.97QCSEE80 pKa = 4.19TKK82 pKa = 9.12EE83 pKa = 4.07CAHH86 pKa = 6.11YY87 pKa = 8.95VHH89 pKa = 7.39HH90 pKa = 5.97YY91 pKa = 7.83QEE93 pKa = 4.48CVEE96 pKa = 4.21RR97 pKa = 11.84VTKK100 pKa = 10.43EE101 pKa = 3.85MEE103 pKa = 3.87EE104 pKa = 3.85PGYY107 pKa = 10.3DD108 pKa = 3.08EE109 pKa = 5.86KK110 pKa = 11.14EE111 pKa = 4.15YY112 pKa = 11.23KK113 pKa = 9.72EE114 pKa = 4.32DD115 pKa = 3.51CVEE118 pKa = 4.17EE119 pKa = 4.55FFHH122 pKa = 6.74LQHH125 pKa = 7.19CINDD129 pKa = 3.77CVAPKK134 pKa = 10.61LFYY137 pKa = 10.45KK138 pKa = 10.84LKK140 pKa = 10.57
Molecular weight: 16.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W1Q7R0|W1Q7R0_OGAPD Protein JSN1 OS=Ogataea parapolymorpha (strain ATCC 26012 / BCRC 20466 / JCM 22074 / NRRL Y-7560 / DL-1) OX=871575 GN=HPODL_02653 PE=4 SV=1
MM1 pKa = 7.49LRR3 pKa = 11.84TIKK6 pKa = 10.35LARR9 pKa = 11.84PISTQRR15 pKa = 11.84EE16 pKa = 3.62RR17 pKa = 11.84RR18 pKa = 11.84FFSTDD23 pKa = 2.86VVSVFEE29 pKa = 4.43SGAEE33 pKa = 4.0LVSAAHH39 pKa = 6.62AASGLPWWAFIPLITVSVRR58 pKa = 11.84TCITLPLAIYY68 pKa = 8.65QRR70 pKa = 11.84RR71 pKa = 11.84GLQKK75 pKa = 10.37QNEE78 pKa = 4.22LRR80 pKa = 11.84PIISAMFPIFKK91 pKa = 10.31LRR93 pKa = 11.84LAARR97 pKa = 11.84AQAAQQSARR106 pKa = 11.84LSKK109 pKa = 10.79SDD111 pKa = 3.64VPIKK115 pKa = 10.24TEE117 pKa = 4.03TEE119 pKa = 3.67QLSANKK125 pKa = 10.0IIVLASKK132 pKa = 10.61EE133 pKa = 3.83RR134 pKa = 11.84MKK136 pKa = 10.57RR137 pKa = 11.84QRR139 pKa = 11.84QIFRR143 pKa = 11.84DD144 pKa = 3.76NGCQSWKK151 pKa = 10.45FLALPALQIPLWITLSQTFRR171 pKa = 11.84VLTGWTNVRR180 pKa = 11.84ATVLDD185 pKa = 3.78PTLSTEE191 pKa = 4.06GLGYY195 pKa = 8.86LTNLTLSDD203 pKa = 4.38PYY205 pKa = 10.85FILPVVLGVTALTNAEE221 pKa = 4.15WNFRR225 pKa = 11.84TADD228 pKa = 3.65LMKK231 pKa = 9.79LTTRR235 pKa = 11.84GVRR238 pKa = 11.84NSLRR242 pKa = 11.84PTAFDD247 pKa = 3.4SVINLTRR254 pKa = 11.84MSICFLVVLSAQAPAALSIYY274 pKa = 9.04WISSNIYY281 pKa = 10.41SLAQNILLNKK291 pKa = 9.34FMPLRR296 pKa = 11.84YY297 pKa = 7.83TPYY300 pKa = 10.67ARR302 pKa = 11.84SAPSTKK308 pKa = 10.23KK309 pKa = 10.45LAGEE313 pKa = 4.19SLVEE317 pKa = 4.26FF318 pKa = 4.95
MM1 pKa = 7.49LRR3 pKa = 11.84TIKK6 pKa = 10.35LARR9 pKa = 11.84PISTQRR15 pKa = 11.84EE16 pKa = 3.62RR17 pKa = 11.84RR18 pKa = 11.84FFSTDD23 pKa = 2.86VVSVFEE29 pKa = 4.43SGAEE33 pKa = 4.0LVSAAHH39 pKa = 6.62AASGLPWWAFIPLITVSVRR58 pKa = 11.84TCITLPLAIYY68 pKa = 8.65QRR70 pKa = 11.84RR71 pKa = 11.84GLQKK75 pKa = 10.37QNEE78 pKa = 4.22LRR80 pKa = 11.84PIISAMFPIFKK91 pKa = 10.31LRR93 pKa = 11.84LAARR97 pKa = 11.84AQAAQQSARR106 pKa = 11.84LSKK109 pKa = 10.79SDD111 pKa = 3.64VPIKK115 pKa = 10.24TEE117 pKa = 4.03TEE119 pKa = 3.67QLSANKK125 pKa = 10.0IIVLASKK132 pKa = 10.61EE133 pKa = 3.83RR134 pKa = 11.84MKK136 pKa = 10.57RR137 pKa = 11.84QRR139 pKa = 11.84QIFRR143 pKa = 11.84DD144 pKa = 3.76NGCQSWKK151 pKa = 10.45FLALPALQIPLWITLSQTFRR171 pKa = 11.84VLTGWTNVRR180 pKa = 11.84ATVLDD185 pKa = 3.78PTLSTEE191 pKa = 4.06GLGYY195 pKa = 8.86LTNLTLSDD203 pKa = 4.38PYY205 pKa = 10.85FILPVVLGVTALTNAEE221 pKa = 4.15WNFRR225 pKa = 11.84TADD228 pKa = 3.65LMKK231 pKa = 9.79LTTRR235 pKa = 11.84GVRR238 pKa = 11.84NSLRR242 pKa = 11.84PTAFDD247 pKa = 3.4SVINLTRR254 pKa = 11.84MSICFLVVLSAQAPAALSIYY274 pKa = 9.04WISSNIYY281 pKa = 10.41SLAQNILLNKK291 pKa = 9.34FMPLRR296 pKa = 11.84YY297 pKa = 7.83TPYY300 pKa = 10.67ARR302 pKa = 11.84SAPSTKK308 pKa = 10.23KK309 pKa = 10.45LAGEE313 pKa = 4.19SLVEE317 pKa = 4.26FF318 pKa = 4.95
Molecular weight: 35.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2502102 |
48 |
4775 |
468.7 |
52.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.533 ± 0.033 | 1.257 ± 0.01 |
5.83 ± 0.023 | 6.872 ± 0.035 |
4.368 ± 0.018 | 5.347 ± 0.031 |
2.122 ± 0.012 | 5.726 ± 0.023 |
6.694 ± 0.03 | 10.158 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.098 ± 0.012 | 4.644 ± 0.019 |
4.616 ± 0.025 | 4.092 ± 0.022 |
4.932 ± 0.026 | 8.328 ± 0.051 |
5.462 ± 0.022 | 6.366 ± 0.024 |
1.073 ± 0.009 | 3.482 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
