
Anaerohalosphaera lusitana
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Phycisphaerae; Sedimentisphaerales; Anaerohalosphaeraceae; Anaerohalosphaera
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
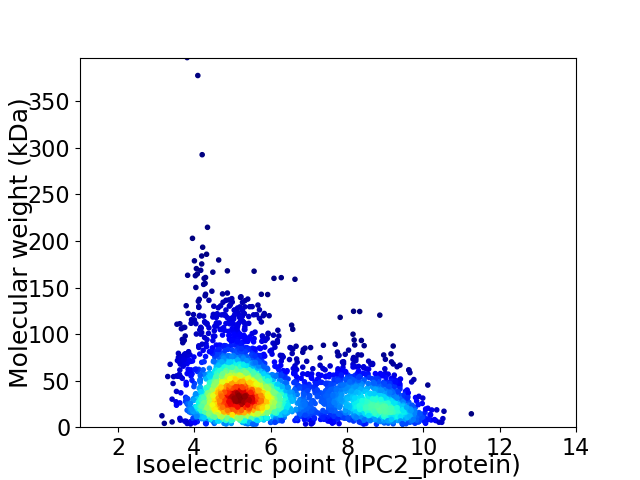
Virtual 2D-PAGE plot for 3413 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U9NN70|A0A1U9NN70_9BACT Uncharacterized protein OS=Anaerohalosphaera lusitana OX=1936003 GN=STSP2_02436 PE=4 SV=1
MM1 pKa = 7.34SFKK4 pKa = 10.68KK5 pKa = 10.79SFVPVVNLGRR15 pKa = 11.84IMRR18 pKa = 11.84RR19 pKa = 11.84CSKK22 pKa = 10.29LFAIIAILLFVSQAGYY38 pKa = 11.14ASLVGHH44 pKa = 7.19WKK46 pKa = 10.47LDD48 pKa = 3.89DD49 pKa = 4.48GSGSTAASAVDD60 pKa = 3.85SPADD64 pKa = 3.77DD65 pKa = 3.91GAIVGTAAWEE75 pKa = 4.37STDD78 pKa = 3.36IPPVPGGNSYY88 pKa = 11.25ALEE91 pKa = 4.06LAAGKK96 pKa = 10.81GIDD99 pKa = 3.84TPVNGIPGAGGRR111 pKa = 11.84TISAWINVDD120 pKa = 3.15PTASLAEE127 pKa = 4.3GGGGIVTWGDD137 pKa = 2.52NWANGLGHH145 pKa = 7.45RR146 pKa = 11.84FTFKK150 pKa = 10.33IDD152 pKa = 3.54DD153 pKa = 3.83SSTGRR158 pKa = 11.84LRR160 pKa = 11.84VEE162 pKa = 3.59IGGGYY167 pKa = 10.41AVGSTSVNDD176 pKa = 4.64GQWHH180 pKa = 5.64HH181 pKa = 6.32VAVTLADD188 pKa = 4.09GQSSVANVKK197 pKa = 9.94FYY199 pKa = 10.67IDD201 pKa = 3.91GEE203 pKa = 4.25VDD205 pKa = 3.81VISDD209 pKa = 4.04SDD211 pKa = 3.6TSNPINSDD219 pKa = 3.2AAPIDD224 pKa = 3.18IGYY227 pKa = 10.11CEE229 pKa = 4.94ALLDD233 pKa = 3.86TWSKK237 pKa = 11.61AMGIEE242 pKa = 4.11GGIDD246 pKa = 3.26DD247 pKa = 4.19VRR249 pKa = 11.84VYY251 pKa = 11.07DD252 pKa = 3.97HH253 pKa = 6.96EE254 pKa = 4.51LSEE257 pKa = 4.98GEE259 pKa = 4.41ILLLCTGSDD268 pKa = 3.33EE269 pKa = 4.65PYY271 pKa = 10.81VYY273 pKa = 10.61GPSPGDD279 pKa = 3.73GEE281 pKa = 5.05DD282 pKa = 3.49HH283 pKa = 6.66VSPDD287 pKa = 4.42AVLSWSAGAVEE298 pKa = 4.41NPDD301 pKa = 3.24YY302 pKa = 11.04DD303 pKa = 4.41VNIGTTMACDD313 pKa = 4.01EE314 pKa = 5.04IISGVSTGSATSYY327 pKa = 10.83VPAAGTLTLGNTYY340 pKa = 9.23FWRR343 pKa = 11.84VDD345 pKa = 3.35VTDD348 pKa = 4.4GGTEE352 pKa = 3.81YY353 pKa = 10.62TGDD356 pKa = 3.03VWSFTVGAGSATDD369 pKa = 3.81PVPADD374 pKa = 3.48GASIVATSAYY384 pKa = 10.62LDD386 pKa = 3.32WTGDD390 pKa = 3.77DD391 pKa = 4.52FVDD394 pKa = 3.98SYY396 pKa = 11.92RR397 pKa = 11.84IMFAPAGEE405 pKa = 4.07QLVNAGEE412 pKa = 4.24YY413 pKa = 10.52AGAPVGLAKK422 pKa = 10.17IARR425 pKa = 11.84AAGMDD430 pKa = 3.84LLEE433 pKa = 5.11AGTYY437 pKa = 9.17DD438 pKa = 3.47WQVVSLDD445 pKa = 3.73ASGEE449 pKa = 4.25MINSGPVWSFSIGTDD464 pKa = 2.07IWVYY468 pKa = 10.32MLGDD472 pKa = 4.0YY473 pKa = 11.08VPVDD477 pKa = 3.39TTVDD481 pKa = 3.49DD482 pKa = 4.16FFSYY486 pKa = 11.14VSTADD491 pKa = 5.25LLGTWTDD498 pKa = 3.71GAANGSNAQATIDD511 pKa = 3.65AMEE514 pKa = 4.4GAVTLSYY521 pKa = 11.67DD522 pKa = 3.52NTTVPYY528 pKa = 10.04QSYY531 pKa = 10.45ISRR534 pKa = 11.84TFASVQDD541 pKa = 3.59WSGEE545 pKa = 4.05QVLSVTMQGDD555 pKa = 3.71SANAGEE561 pKa = 4.3TLFVGLSDD569 pKa = 4.04GTDD572 pKa = 3.43SADD575 pKa = 4.46LALQSEE581 pKa = 4.71KK582 pKa = 10.85LVSNDD587 pKa = 2.76EE588 pKa = 4.13ATTITLALSEE598 pKa = 4.16FAAAGVDD605 pKa = 3.69LANVSEE611 pKa = 4.02MRR613 pKa = 11.84IGTGDD618 pKa = 3.43GSTGGAAGSIVVYY631 pKa = 10.73DD632 pKa = 4.2VMIHH636 pKa = 6.11PAGSEE641 pKa = 4.13EE642 pKa = 4.04ITEE645 pKa = 4.14PTADD649 pKa = 3.8LDD651 pKa = 3.84GDD653 pKa = 4.1FVVDD657 pKa = 5.01LGDD660 pKa = 3.19IALFAEE666 pKa = 5.57GWAMAEE672 pKa = 4.17YY673 pKa = 9.22MVSASGSEE681 pKa = 4.06PAGLRR686 pKa = 11.84AEE688 pKa = 4.2YY689 pKa = 10.31RR690 pKa = 11.84FEE692 pKa = 3.92EE693 pKa = 4.2LGGYY697 pKa = 8.59VVGDD701 pKa = 3.54SSGNGLDD708 pKa = 4.53GSIQRR713 pKa = 11.84DD714 pKa = 3.58LTGVWNVDD722 pKa = 3.44GFDD725 pKa = 3.91GGCVKK730 pKa = 9.96LTDD733 pKa = 4.07GSSVVLPASVFDD745 pKa = 4.24GVDD748 pKa = 3.3GFTMSFWMSQTTSAGVGMQAFTVDD772 pKa = 3.74SVGLDD777 pKa = 3.14ASYY780 pKa = 11.4EE781 pKa = 4.0LTNTGFYY788 pKa = 10.58DD789 pKa = 4.66GSWHH793 pKa = 7.11HH794 pKa = 6.2IAIVNDD800 pKa = 4.76AINARR805 pKa = 11.84VEE807 pKa = 4.28LYY809 pKa = 11.43VNGLLVAGGDD819 pKa = 3.4AGSTAEE825 pKa = 4.28AASEE829 pKa = 4.08TVIEE833 pKa = 4.95KK834 pKa = 10.45ISDD837 pKa = 3.46NAFGSTVDD845 pKa = 4.38KK846 pKa = 10.49IDD848 pKa = 3.55SVSVYY853 pKa = 10.67SRR855 pKa = 11.84ALDD858 pKa = 3.23HH859 pKa = 7.08SEE861 pKa = 3.46IVRR864 pKa = 11.84LYY866 pKa = 9.48GGAGASVMQPVNGSVTVSDD885 pKa = 4.2LNGDD889 pKa = 3.66GRR891 pKa = 11.84VDD893 pKa = 3.72LEE895 pKa = 5.44DD896 pKa = 3.56YY897 pKa = 11.45AEE899 pKa = 5.35LIGSMIQRR907 pKa = 11.84LL908 pKa = 3.59
MM1 pKa = 7.34SFKK4 pKa = 10.68KK5 pKa = 10.79SFVPVVNLGRR15 pKa = 11.84IMRR18 pKa = 11.84RR19 pKa = 11.84CSKK22 pKa = 10.29LFAIIAILLFVSQAGYY38 pKa = 11.14ASLVGHH44 pKa = 7.19WKK46 pKa = 10.47LDD48 pKa = 3.89DD49 pKa = 4.48GSGSTAASAVDD60 pKa = 3.85SPADD64 pKa = 3.77DD65 pKa = 3.91GAIVGTAAWEE75 pKa = 4.37STDD78 pKa = 3.36IPPVPGGNSYY88 pKa = 11.25ALEE91 pKa = 4.06LAAGKK96 pKa = 10.81GIDD99 pKa = 3.84TPVNGIPGAGGRR111 pKa = 11.84TISAWINVDD120 pKa = 3.15PTASLAEE127 pKa = 4.3GGGGIVTWGDD137 pKa = 2.52NWANGLGHH145 pKa = 7.45RR146 pKa = 11.84FTFKK150 pKa = 10.33IDD152 pKa = 3.54DD153 pKa = 3.83SSTGRR158 pKa = 11.84LRR160 pKa = 11.84VEE162 pKa = 3.59IGGGYY167 pKa = 10.41AVGSTSVNDD176 pKa = 4.64GQWHH180 pKa = 5.64HH181 pKa = 6.32VAVTLADD188 pKa = 4.09GQSSVANVKK197 pKa = 9.94FYY199 pKa = 10.67IDD201 pKa = 3.91GEE203 pKa = 4.25VDD205 pKa = 3.81VISDD209 pKa = 4.04SDD211 pKa = 3.6TSNPINSDD219 pKa = 3.2AAPIDD224 pKa = 3.18IGYY227 pKa = 10.11CEE229 pKa = 4.94ALLDD233 pKa = 3.86TWSKK237 pKa = 11.61AMGIEE242 pKa = 4.11GGIDD246 pKa = 3.26DD247 pKa = 4.19VRR249 pKa = 11.84VYY251 pKa = 11.07DD252 pKa = 3.97HH253 pKa = 6.96EE254 pKa = 4.51LSEE257 pKa = 4.98GEE259 pKa = 4.41ILLLCTGSDD268 pKa = 3.33EE269 pKa = 4.65PYY271 pKa = 10.81VYY273 pKa = 10.61GPSPGDD279 pKa = 3.73GEE281 pKa = 5.05DD282 pKa = 3.49HH283 pKa = 6.66VSPDD287 pKa = 4.42AVLSWSAGAVEE298 pKa = 4.41NPDD301 pKa = 3.24YY302 pKa = 11.04DD303 pKa = 4.41VNIGTTMACDD313 pKa = 4.01EE314 pKa = 5.04IISGVSTGSATSYY327 pKa = 10.83VPAAGTLTLGNTYY340 pKa = 9.23FWRR343 pKa = 11.84VDD345 pKa = 3.35VTDD348 pKa = 4.4GGTEE352 pKa = 3.81YY353 pKa = 10.62TGDD356 pKa = 3.03VWSFTVGAGSATDD369 pKa = 3.81PVPADD374 pKa = 3.48GASIVATSAYY384 pKa = 10.62LDD386 pKa = 3.32WTGDD390 pKa = 3.77DD391 pKa = 4.52FVDD394 pKa = 3.98SYY396 pKa = 11.92RR397 pKa = 11.84IMFAPAGEE405 pKa = 4.07QLVNAGEE412 pKa = 4.24YY413 pKa = 10.52AGAPVGLAKK422 pKa = 10.17IARR425 pKa = 11.84AAGMDD430 pKa = 3.84LLEE433 pKa = 5.11AGTYY437 pKa = 9.17DD438 pKa = 3.47WQVVSLDD445 pKa = 3.73ASGEE449 pKa = 4.25MINSGPVWSFSIGTDD464 pKa = 2.07IWVYY468 pKa = 10.32MLGDD472 pKa = 4.0YY473 pKa = 11.08VPVDD477 pKa = 3.39TTVDD481 pKa = 3.49DD482 pKa = 4.16FFSYY486 pKa = 11.14VSTADD491 pKa = 5.25LLGTWTDD498 pKa = 3.71GAANGSNAQATIDD511 pKa = 3.65AMEE514 pKa = 4.4GAVTLSYY521 pKa = 11.67DD522 pKa = 3.52NTTVPYY528 pKa = 10.04QSYY531 pKa = 10.45ISRR534 pKa = 11.84TFASVQDD541 pKa = 3.59WSGEE545 pKa = 4.05QVLSVTMQGDD555 pKa = 3.71SANAGEE561 pKa = 4.3TLFVGLSDD569 pKa = 4.04GTDD572 pKa = 3.43SADD575 pKa = 4.46LALQSEE581 pKa = 4.71KK582 pKa = 10.85LVSNDD587 pKa = 2.76EE588 pKa = 4.13ATTITLALSEE598 pKa = 4.16FAAAGVDD605 pKa = 3.69LANVSEE611 pKa = 4.02MRR613 pKa = 11.84IGTGDD618 pKa = 3.43GSTGGAAGSIVVYY631 pKa = 10.73DD632 pKa = 4.2VMIHH636 pKa = 6.11PAGSEE641 pKa = 4.13EE642 pKa = 4.04ITEE645 pKa = 4.14PTADD649 pKa = 3.8LDD651 pKa = 3.84GDD653 pKa = 4.1FVVDD657 pKa = 5.01LGDD660 pKa = 3.19IALFAEE666 pKa = 5.57GWAMAEE672 pKa = 4.17YY673 pKa = 9.22MVSASGSEE681 pKa = 4.06PAGLRR686 pKa = 11.84AEE688 pKa = 4.2YY689 pKa = 10.31RR690 pKa = 11.84FEE692 pKa = 3.92EE693 pKa = 4.2LGGYY697 pKa = 8.59VVGDD701 pKa = 3.54SSGNGLDD708 pKa = 4.53GSIQRR713 pKa = 11.84DD714 pKa = 3.58LTGVWNVDD722 pKa = 3.44GFDD725 pKa = 3.91GGCVKK730 pKa = 9.96LTDD733 pKa = 4.07GSSVVLPASVFDD745 pKa = 4.24GVDD748 pKa = 3.3GFTMSFWMSQTTSAGVGMQAFTVDD772 pKa = 3.74SVGLDD777 pKa = 3.14ASYY780 pKa = 11.4EE781 pKa = 4.0LTNTGFYY788 pKa = 10.58DD789 pKa = 4.66GSWHH793 pKa = 7.11HH794 pKa = 6.2IAIVNDD800 pKa = 4.76AINARR805 pKa = 11.84VEE807 pKa = 4.28LYY809 pKa = 11.43VNGLLVAGGDD819 pKa = 3.4AGSTAEE825 pKa = 4.28AASEE829 pKa = 4.08TVIEE833 pKa = 4.95KK834 pKa = 10.45ISDD837 pKa = 3.46NAFGSTVDD845 pKa = 4.38KK846 pKa = 10.49IDD848 pKa = 3.55SVSVYY853 pKa = 10.67SRR855 pKa = 11.84ALDD858 pKa = 3.23HH859 pKa = 7.08SEE861 pKa = 3.46IVRR864 pKa = 11.84LYY866 pKa = 9.48GGAGASVMQPVNGSVTVSDD885 pKa = 4.2LNGDD889 pKa = 3.66GRR891 pKa = 11.84VDD893 pKa = 3.72LEE895 pKa = 5.44DD896 pKa = 3.56YY897 pKa = 11.45AEE899 pKa = 5.35LIGSMIQRR907 pKa = 11.84LL908 pKa = 3.59
Molecular weight: 94.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U9NLP7|A0A1U9NLP7_9BACT Replication-associated recombination protein A OS=Anaerohalosphaera lusitana OX=1936003 GN=rarA PE=4 SV=1
MM1 pKa = 7.51PRR3 pKa = 11.84PVRR6 pKa = 11.84CRR8 pKa = 11.84RR9 pKa = 11.84VGMTPKK15 pKa = 9.47CTYY18 pKa = 9.83FKK20 pKa = 10.17PQGVRR25 pKa = 11.84LSALEE30 pKa = 4.13VVGLTVDD37 pKa = 3.13EE38 pKa = 4.83LEE40 pKa = 4.35AVRR43 pKa = 11.84LADD46 pKa = 5.51LEE48 pKa = 4.23GHH50 pKa = 5.42YY51 pKa = 8.23QQCAAEE57 pKa = 4.23KK58 pKa = 9.48MGVSRR63 pKa = 11.84QTFGRR68 pKa = 11.84IIEE71 pKa = 4.31SAHH74 pKa = 6.11KK75 pKa = 10.09KK76 pKa = 8.9IAEE79 pKa = 3.93ALVNGKK85 pKa = 9.28ALSIEE90 pKa = 4.21GGVVNLPEE98 pKa = 4.06EE99 pKa = 4.12ARR101 pKa = 11.84GRR103 pKa = 11.84GQGRR107 pKa = 11.84GRR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84GGGPGRR118 pKa = 11.84GRR120 pKa = 11.84CGQQ123 pKa = 3.26
MM1 pKa = 7.51PRR3 pKa = 11.84PVRR6 pKa = 11.84CRR8 pKa = 11.84RR9 pKa = 11.84VGMTPKK15 pKa = 9.47CTYY18 pKa = 9.83FKK20 pKa = 10.17PQGVRR25 pKa = 11.84LSALEE30 pKa = 4.13VVGLTVDD37 pKa = 3.13EE38 pKa = 4.83LEE40 pKa = 4.35AVRR43 pKa = 11.84LADD46 pKa = 5.51LEE48 pKa = 4.23GHH50 pKa = 5.42YY51 pKa = 8.23QQCAAEE57 pKa = 4.23KK58 pKa = 9.48MGVSRR63 pKa = 11.84QTFGRR68 pKa = 11.84IIEE71 pKa = 4.31SAHH74 pKa = 6.11KK75 pKa = 10.09KK76 pKa = 8.9IAEE79 pKa = 3.93ALVNGKK85 pKa = 9.28ALSIEE90 pKa = 4.21GGVVNLPEE98 pKa = 4.06EE99 pKa = 4.12ARR101 pKa = 11.84GRR103 pKa = 11.84GQGRR107 pKa = 11.84GRR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84GGGPGRR118 pKa = 11.84GRR120 pKa = 11.84CGQQ123 pKa = 3.26
Molecular weight: 13.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1238531 |
29 |
3724 |
362.9 |
40.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.512 ± 0.045 | 1.336 ± 0.017 |
6.493 ± 0.037 | 6.776 ± 0.049 |
3.945 ± 0.026 | 7.874 ± 0.047 |
1.931 ± 0.02 | 5.992 ± 0.036 |
5.544 ± 0.048 | 8.634 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.575 ± 0.021 | 3.945 ± 0.029 |
4.196 ± 0.029 | 3.457 ± 0.028 |
5.258 ± 0.036 | 6.084 ± 0.037 |
5.473 ± 0.045 | 7.163 ± 0.035 |
1.462 ± 0.019 | 3.35 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
