
Sewage-associated circular DNA virus-25
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
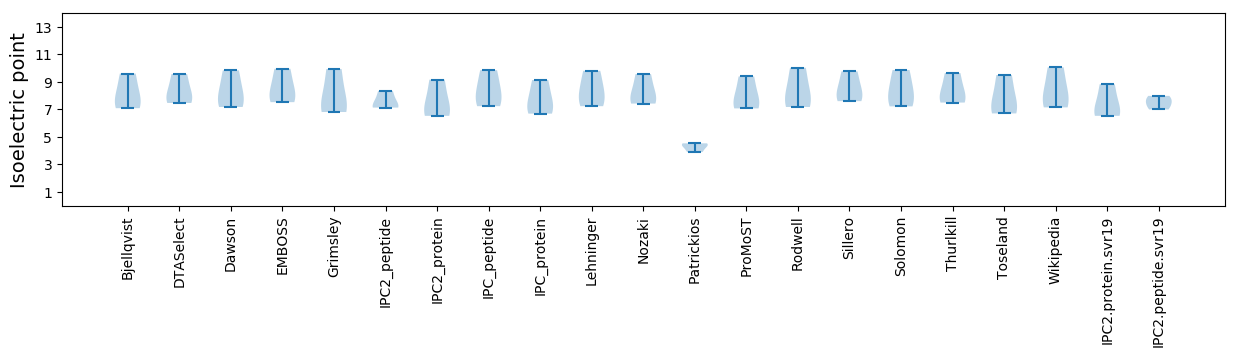
Average proteome isoelectric point is 7.43
Get precalculated fractions of proteins
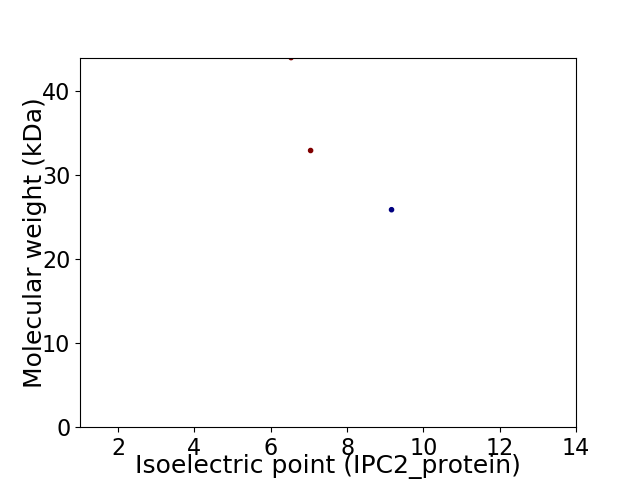
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UH84|A0A0B4UH84_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-25 OX=1592092 PE=4 SV=1
MM1 pKa = 7.42SSSSRR6 pKa = 11.84WVFTVNNPGAWRR18 pKa = 11.84PHH20 pKa = 6.1YY21 pKa = 10.46DD22 pKa = 3.07VTAMQFLVFQMEE34 pKa = 4.25QGASGTPHH42 pKa = 5.25IQGYY46 pKa = 8.29VRR48 pKa = 11.84FTHH51 pKa = 6.12PKK53 pKa = 9.03KK54 pKa = 10.92LEE56 pKa = 3.82AAKK59 pKa = 10.25RR60 pKa = 11.84ALVCEE65 pKa = 4.26EE66 pKa = 3.61AHH68 pKa = 6.33MEE70 pKa = 3.92IANGNEE76 pKa = 4.16GQCIDD81 pKa = 3.94YY82 pKa = 8.27CTKK85 pKa = 9.99EE86 pKa = 4.04DD87 pKa = 3.71TRR89 pKa = 11.84IGGPWVYY96 pKa = 9.41GTQKK100 pKa = 10.3RR101 pKa = 11.84DD102 pKa = 3.15AGRR105 pKa = 11.84KK106 pKa = 8.66GSRR109 pKa = 11.84SDD111 pKa = 3.5LQAVAEE117 pKa = 4.27AAQKK121 pKa = 10.28PGVSMRR127 pKa = 11.84QLARR131 pKa = 11.84EE132 pKa = 4.53FPAQVIKK139 pKa = 10.84YY140 pKa = 7.55SQQIEE145 pKa = 4.35RR146 pKa = 11.84YY147 pKa = 7.03MALVEE152 pKa = 4.31DD153 pKa = 4.46EE154 pKa = 4.63VPIEE158 pKa = 3.75RR159 pKa = 11.84DD160 pKa = 3.05IFVHH164 pKa = 6.17VIWGPTGTGKK174 pKa = 7.06THH176 pKa = 6.89RR177 pKa = 11.84VRR179 pKa = 11.84HH180 pKa = 6.33GIPQRR185 pKa = 11.84DD186 pKa = 3.67LYY188 pKa = 10.71VVNGKK193 pKa = 9.52GRR195 pKa = 11.84GCFDD199 pKa = 3.06QYY201 pKa = 11.62AGQKK205 pKa = 10.62ALVLEE210 pKa = 4.31EE211 pKa = 4.55FEE213 pKa = 4.52PNEE216 pKa = 3.6WSINEE221 pKa = 4.16VKK223 pKa = 10.01TLLDD227 pKa = 3.16KK228 pKa = 7.62WTCPLNARR236 pKa = 11.84YY237 pKa = 9.95SNKK240 pKa = 7.53TARR243 pKa = 11.84WTQVFITSNSDD254 pKa = 2.95PMSWYY259 pKa = 10.16QLWLPADD266 pKa = 3.92LAAFQRR272 pKa = 11.84RR273 pKa = 11.84LTRR276 pKa = 11.84ITHH279 pKa = 4.65VLSRR283 pKa = 11.84EE284 pKa = 4.03QEE286 pKa = 4.14VEE288 pKa = 4.37LFPLPAVALPPPSPAVAPSPIAARR312 pKa = 11.84IAARR316 pKa = 11.84PCWSGAMDD324 pKa = 5.29ADD326 pKa = 5.0LPQPDD331 pKa = 4.38QVTSADD337 pKa = 3.76PLRR340 pKa = 11.84AAGTSNAPAIPTPRR354 pKa = 11.84LPTTPTPDD362 pKa = 3.77PLNVQDD368 pKa = 4.25IDD370 pKa = 4.33GIGAITAPAPVTRR383 pKa = 11.84TTSVDD388 pKa = 3.35LQAFFSNNN396 pKa = 2.84
MM1 pKa = 7.42SSSSRR6 pKa = 11.84WVFTVNNPGAWRR18 pKa = 11.84PHH20 pKa = 6.1YY21 pKa = 10.46DD22 pKa = 3.07VTAMQFLVFQMEE34 pKa = 4.25QGASGTPHH42 pKa = 5.25IQGYY46 pKa = 8.29VRR48 pKa = 11.84FTHH51 pKa = 6.12PKK53 pKa = 9.03KK54 pKa = 10.92LEE56 pKa = 3.82AAKK59 pKa = 10.25RR60 pKa = 11.84ALVCEE65 pKa = 4.26EE66 pKa = 3.61AHH68 pKa = 6.33MEE70 pKa = 3.92IANGNEE76 pKa = 4.16GQCIDD81 pKa = 3.94YY82 pKa = 8.27CTKK85 pKa = 9.99EE86 pKa = 4.04DD87 pKa = 3.71TRR89 pKa = 11.84IGGPWVYY96 pKa = 9.41GTQKK100 pKa = 10.3RR101 pKa = 11.84DD102 pKa = 3.15AGRR105 pKa = 11.84KK106 pKa = 8.66GSRR109 pKa = 11.84SDD111 pKa = 3.5LQAVAEE117 pKa = 4.27AAQKK121 pKa = 10.28PGVSMRR127 pKa = 11.84QLARR131 pKa = 11.84EE132 pKa = 4.53FPAQVIKK139 pKa = 10.84YY140 pKa = 7.55SQQIEE145 pKa = 4.35RR146 pKa = 11.84YY147 pKa = 7.03MALVEE152 pKa = 4.31DD153 pKa = 4.46EE154 pKa = 4.63VPIEE158 pKa = 3.75RR159 pKa = 11.84DD160 pKa = 3.05IFVHH164 pKa = 6.17VIWGPTGTGKK174 pKa = 7.06THH176 pKa = 6.89RR177 pKa = 11.84VRR179 pKa = 11.84HH180 pKa = 6.33GIPQRR185 pKa = 11.84DD186 pKa = 3.67LYY188 pKa = 10.71VVNGKK193 pKa = 9.52GRR195 pKa = 11.84GCFDD199 pKa = 3.06QYY201 pKa = 11.62AGQKK205 pKa = 10.62ALVLEE210 pKa = 4.31EE211 pKa = 4.55FEE213 pKa = 4.52PNEE216 pKa = 3.6WSINEE221 pKa = 4.16VKK223 pKa = 10.01TLLDD227 pKa = 3.16KK228 pKa = 7.62WTCPLNARR236 pKa = 11.84YY237 pKa = 9.95SNKK240 pKa = 7.53TARR243 pKa = 11.84WTQVFITSNSDD254 pKa = 2.95PMSWYY259 pKa = 10.16QLWLPADD266 pKa = 3.92LAAFQRR272 pKa = 11.84RR273 pKa = 11.84LTRR276 pKa = 11.84ITHH279 pKa = 4.65VLSRR283 pKa = 11.84EE284 pKa = 4.03QEE286 pKa = 4.14VEE288 pKa = 4.37LFPLPAVALPPPSPAVAPSPIAARR312 pKa = 11.84IAARR316 pKa = 11.84PCWSGAMDD324 pKa = 5.29ADD326 pKa = 5.0LPQPDD331 pKa = 4.38QVTSADD337 pKa = 3.76PLRR340 pKa = 11.84AAGTSNAPAIPTPRR354 pKa = 11.84LPTTPTPDD362 pKa = 3.77PLNVQDD368 pKa = 4.25IDD370 pKa = 4.33GIGAITAPAPVTRR383 pKa = 11.84TTSVDD388 pKa = 3.35LQAFFSNNN396 pKa = 2.84
Molecular weight: 43.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGY5|A0A0B4UGY5_9VIRU Uncharacterized protein OS=Sewage-associated circular DNA virus-25 OX=1592092 PE=4 SV=1
MM1 pKa = 7.46ARR3 pKa = 11.84YY4 pKa = 8.62KK5 pKa = 10.55RR6 pKa = 11.84PSILSRR12 pKa = 11.84LKK14 pKa = 7.96TTKK17 pKa = 9.19KK18 pKa = 4.95TTRR21 pKa = 11.84GKK23 pKa = 8.0RR24 pKa = 11.84TKK26 pKa = 10.46YY27 pKa = 10.31RR28 pKa = 11.84LGSLGGAPKK37 pKa = 10.8AEE39 pKa = 4.02VKK41 pKa = 11.01SFDD44 pKa = 3.62TYY46 pKa = 11.45VLSNATGLPHH56 pKa = 7.03EE57 pKa = 5.35DD58 pKa = 2.88SAGFAEE64 pKa = 4.56PTSNFAGYY72 pKa = 7.93TCLNEE77 pKa = 4.31VSQGAAFYY85 pKa = 11.15ARR87 pKa = 11.84IGSKK91 pKa = 10.14ISIKK95 pKa = 10.35SIQVDD100 pKa = 4.34FDD102 pKa = 5.12LYY104 pKa = 10.06TLSITTTAGTARR116 pKa = 11.84CILIYY121 pKa = 10.56DD122 pKa = 4.11RR123 pKa = 11.84QTNGAAPSFVDD134 pKa = 3.78MFNAYY139 pKa = 8.4PAAINAFGTGINMTNRR155 pKa = 11.84SRR157 pKa = 11.84FSIIRR162 pKa = 11.84DD163 pKa = 3.15KK164 pKa = 11.26YY165 pKa = 9.12YY166 pKa = 10.56EE167 pKa = 4.28FGPGEE172 pKa = 4.26STTKK176 pKa = 9.95HH177 pKa = 3.55VHH179 pKa = 5.48MYY181 pKa = 11.13AKK183 pKa = 10.3GRR185 pKa = 11.84WDD187 pKa = 3.51TEE189 pKa = 4.63YY190 pKa = 11.12KK191 pKa = 10.67SSTPSVGDD199 pKa = 3.18ITTGAIYY206 pKa = 10.41LSVISNISLAYY217 pKa = 10.14SPGKK221 pKa = 9.83ICMNEE226 pKa = 2.89ITARR230 pKa = 11.84IRR232 pKa = 11.84YY233 pKa = 9.2LDD235 pKa = 3.32
MM1 pKa = 7.46ARR3 pKa = 11.84YY4 pKa = 8.62KK5 pKa = 10.55RR6 pKa = 11.84PSILSRR12 pKa = 11.84LKK14 pKa = 7.96TTKK17 pKa = 9.19KK18 pKa = 4.95TTRR21 pKa = 11.84GKK23 pKa = 8.0RR24 pKa = 11.84TKK26 pKa = 10.46YY27 pKa = 10.31RR28 pKa = 11.84LGSLGGAPKK37 pKa = 10.8AEE39 pKa = 4.02VKK41 pKa = 11.01SFDD44 pKa = 3.62TYY46 pKa = 11.45VLSNATGLPHH56 pKa = 7.03EE57 pKa = 5.35DD58 pKa = 2.88SAGFAEE64 pKa = 4.56PTSNFAGYY72 pKa = 7.93TCLNEE77 pKa = 4.31VSQGAAFYY85 pKa = 11.15ARR87 pKa = 11.84IGSKK91 pKa = 10.14ISIKK95 pKa = 10.35SIQVDD100 pKa = 4.34FDD102 pKa = 5.12LYY104 pKa = 10.06TLSITTTAGTARR116 pKa = 11.84CILIYY121 pKa = 10.56DD122 pKa = 4.11RR123 pKa = 11.84QTNGAAPSFVDD134 pKa = 3.78MFNAYY139 pKa = 8.4PAAINAFGTGINMTNRR155 pKa = 11.84SRR157 pKa = 11.84FSIIRR162 pKa = 11.84DD163 pKa = 3.15KK164 pKa = 11.26YY165 pKa = 9.12YY166 pKa = 10.56EE167 pKa = 4.28FGPGEE172 pKa = 4.26STTKK176 pKa = 9.95HH177 pKa = 3.55VHH179 pKa = 5.48MYY181 pKa = 11.13AKK183 pKa = 10.3GRR185 pKa = 11.84WDD187 pKa = 3.51TEE189 pKa = 4.63YY190 pKa = 11.12KK191 pKa = 10.67SSTPSVGDD199 pKa = 3.18ITTGAIYY206 pKa = 10.41LSVISNISLAYY217 pKa = 10.14SPGKK221 pKa = 9.83ICMNEE226 pKa = 2.89ITARR230 pKa = 11.84IRR232 pKa = 11.84YY233 pKa = 9.2LDD235 pKa = 3.32
Molecular weight: 25.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
916 |
235 |
396 |
305.3 |
34.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.515 ± 1.55 | 1.201 ± 0.219 |
4.803 ± 0.189 | 4.803 ± 0.476 |
4.148 ± 0.528 | 5.459 ± 1.325 |
1.856 ± 0.199 | 7.205 ± 1.366 |
4.913 ± 0.738 | 6.987 ± 0.906 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.528 ± 0.493 | 4.694 ± 0.851 |
6.769 ± 1.193 | 3.603 ± 1.18 |
6.114 ± 0.518 | 7.424 ± 1.036 |
7.969 ± 0.795 | 5.786 ± 0.856 |
1.419 ± 0.586 | 4.803 ± 1.217 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
