
Streptomonospora alba
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae;
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
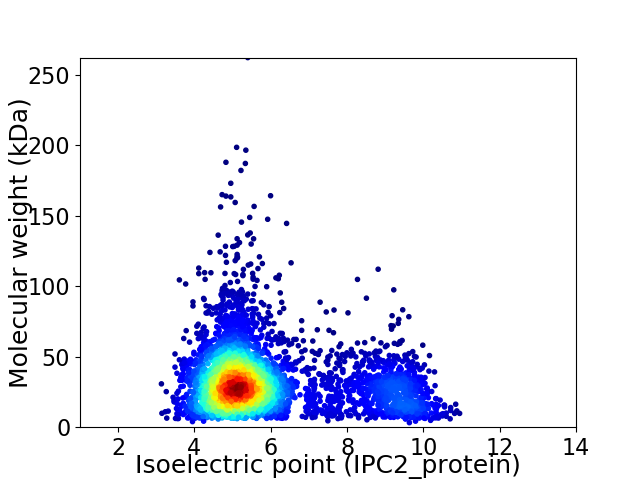
Virtual 2D-PAGE plot for 4307 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C2FEN2|A0A0C2FEN2_9ACTN Uncharacterized protein OS=Streptomonospora alba OX=183763 GN=LP52_18155 PE=4 SV=1
MM1 pKa = 7.42SPSSSRR7 pKa = 11.84SRR9 pKa = 11.84FAAGAVTAASAIALSMLAGAPGAHH33 pKa = 6.75AAEE36 pKa = 4.69LVTNGGFDD44 pKa = 3.45NGTTGWWNTEE54 pKa = 3.64NAPAEE59 pKa = 4.34VADD62 pKa = 4.29GRR64 pKa = 11.84LCADD68 pKa = 3.72VEE70 pKa = 4.5GGTANAWDD78 pKa = 5.1AIVGQDD84 pKa = 4.84GIALTSGEE92 pKa = 4.4SYY94 pKa = 10.96EE95 pKa = 4.3FTFTASASEE104 pKa = 4.31GTTVRR109 pKa = 11.84ALIQEE114 pKa = 3.99PTEE117 pKa = 4.43PYY119 pKa = 9.73TAFLTANPALTSEE132 pKa = 4.5TQTFSHH138 pKa = 7.02TFTADD143 pKa = 2.85ATLDD147 pKa = 3.63DD148 pKa = 4.44AQLVFQLGGEE158 pKa = 4.14AEE160 pKa = 4.08AWTFCLDD167 pKa = 4.62DD168 pKa = 4.65VSLQDD173 pKa = 3.54GAEE176 pKa = 4.08PPVYY180 pKa = 10.25EE181 pKa = 5.39PDD183 pKa = 2.95TGPPVKK189 pKa = 10.39VNQVGYY195 pKa = 10.69LPDD198 pKa = 3.93GPKK201 pKa = 10.39NATLVTDD208 pKa = 4.1ATSAQPWTLRR218 pKa = 11.84DD219 pKa = 3.23ADD221 pKa = 3.87GAAVAEE227 pKa = 4.93GQTSPQGTEE236 pKa = 3.55QSSGQNVHH244 pKa = 6.84SIDD247 pKa = 3.67FSDD250 pKa = 3.88YY251 pKa = 8.24TEE253 pKa = 4.61KK254 pKa = 11.12GSGYY258 pKa = 8.19TLAVGDD264 pKa = 4.52DD265 pKa = 3.59VSHH268 pKa = 7.21PFDD271 pKa = 3.73ISGSAYY277 pKa = 9.65KK278 pKa = 10.41DD279 pKa = 3.35LRR281 pKa = 11.84TDD283 pKa = 3.43SLSLYY288 pKa = 8.96YY289 pKa = 9.36PQRR292 pKa = 11.84SGIEE296 pKa = 3.81IDD298 pKa = 3.92DD299 pKa = 4.04SLMPGYY305 pKa = 10.28GRR307 pKa = 11.84AAGHH311 pKa = 6.28VGVEE315 pKa = 4.23PNQGDD320 pKa = 3.96LSVPCAPDD328 pKa = 3.18SGCDD332 pKa = 3.53YY333 pKa = 10.94EE334 pKa = 6.2LDD336 pKa = 4.33VSGGWYY342 pKa = 10.02DD343 pKa = 3.88AGDD346 pKa = 3.49HH347 pKa = 6.07GKK349 pKa = 10.25YY350 pKa = 9.61VVNGGISASQVMSVWEE366 pKa = 4.04RR367 pKa = 11.84TTYY370 pKa = 11.23ADD372 pKa = 3.19SASPDD377 pKa = 3.35RR378 pKa = 11.84LGDD381 pKa = 3.3GTLPVPEE388 pKa = 4.72QGNDD392 pKa = 3.28VPDD395 pKa = 3.92VLDD398 pKa = 3.64EE399 pKa = 4.8ARR401 pKa = 11.84WEE403 pKa = 4.06MDD405 pKa = 3.19FLMKK409 pKa = 9.91MQVPEE414 pKa = 4.27GEE416 pKa = 4.34EE417 pKa = 3.79LAGMAHH423 pKa = 6.57HH424 pKa = 6.95KK425 pKa = 9.3MHH427 pKa = 7.0DD428 pKa = 4.03EE429 pKa = 4.01EE430 pKa = 4.82WTGLPLMPADD440 pKa = 4.69DD441 pKa = 4.28PQPRR445 pKa = 11.84YY446 pKa = 9.43LQPPSTAATLNLAATGAQCARR467 pKa = 11.84VFEE470 pKa = 5.01SYY472 pKa = 10.87DD473 pKa = 3.25SEE475 pKa = 4.31FADD478 pKa = 3.59RR479 pKa = 11.84CLEE482 pKa = 4.09TAEE485 pKa = 4.9KK486 pKa = 10.02AWQAAQEE493 pKa = 4.29NPDD496 pKa = 3.68VLADD500 pKa = 3.78PEE502 pKa = 4.31NGVGGGAYY510 pKa = 10.23SDD512 pKa = 5.21DD513 pKa = 3.83DD514 pKa = 4.72VSDD517 pKa = 3.53EE518 pKa = 4.94FYY520 pKa = 10.34WAAAEE525 pKa = 4.28LFITTGEE532 pKa = 3.91QSYY535 pKa = 11.18EE536 pKa = 4.06DD537 pKa = 3.6AVLDD541 pKa = 4.34HH542 pKa = 6.89EE543 pKa = 4.93LHH545 pKa = 6.05TEE547 pKa = 4.07DD548 pKa = 4.55VFTAGGLAWGAVAPLGRR565 pKa = 11.84MNLALVPNKK574 pKa = 10.33LDD576 pKa = 3.68DD577 pKa = 3.94RR578 pKa = 11.84DD579 pKa = 3.78RR580 pKa = 11.84VVDD583 pKa = 4.07SVVAGADD590 pKa = 3.51EE591 pKa = 4.16YY592 pKa = 11.71LDD594 pKa = 3.53TLRR597 pKa = 11.84NHH599 pKa = 7.2AFGLPYY605 pKa = 10.52APPDD609 pKa = 3.52NVFVWGSNSQVLNNMVVMTAAYY631 pKa = 10.33DD632 pKa = 3.72LTGDD636 pKa = 3.57GAYY639 pKa = 10.07RR640 pKa = 11.84DD641 pKa = 4.1GVLEE645 pKa = 4.21GMDD648 pKa = 3.88YY649 pKa = 10.92ILGRR653 pKa = 11.84NALGQSYY660 pKa = 7.56VTGYY664 pKa = 11.12GEE666 pKa = 3.97QFSKK670 pKa = 10.56NQHH673 pKa = 4.93SRR675 pKa = 11.84WYY677 pKa = 10.58ANQLDD682 pKa = 4.36ADD684 pKa = 4.48LPNPPEE690 pKa = 4.32GTVSGGPNSQTSTWDD705 pKa = 3.09PVAQDD710 pKa = 4.0KK711 pKa = 11.73LEE713 pKa = 4.31GCAPQFCYY721 pKa = 10.09IDD723 pKa = 5.73DD724 pKa = 4.03IEE726 pKa = 4.27SWATNEE732 pKa = 5.0LTVNWNAPLAIVSSFIADD750 pKa = 3.36QTGEE754 pKa = 4.23GDD756 pKa = 3.84PPAEE760 pKa = 5.1DD761 pKa = 3.48EE762 pKa = 4.53TAPTAPGKK770 pKa = 9.16PQVSGVTSSQAEE782 pKa = 4.45LTWEE786 pKa = 4.06ASTDD790 pKa = 3.44EE791 pKa = 4.73GGSGVEE797 pKa = 4.61GYY799 pKa = 10.24DD800 pKa = 3.44VYY802 pKa = 11.57AGTGDD807 pKa = 3.75GAQKK811 pKa = 10.49VGSASGPSFTLTGLEE826 pKa = 4.28SEE828 pKa = 4.67TEE830 pKa = 4.24YY831 pKa = 10.5TVHH834 pKa = 6.25VVARR838 pKa = 11.84DD839 pKa = 3.52GAGNRR844 pKa = 11.84SEE846 pKa = 4.81PGPNAAFTTEE856 pKa = 3.69AGGTGGEE863 pKa = 4.24AACEE867 pKa = 3.82VDD869 pKa = 4.71YY870 pKa = 10.42STNDD874 pKa = 2.96WNGGFTASVSVVNTGDD890 pKa = 3.29TPIRR894 pKa = 11.84DD895 pKa = 3.38WEE897 pKa = 4.37LGFEE901 pKa = 4.7FGAGQEE907 pKa = 4.24ITHH910 pKa = 6.43GWNAEE915 pKa = 3.61WSQDD919 pKa = 3.14GSAVTASGLSWNSDD933 pKa = 3.7LAPGSSVSAGFNGTAQDD950 pKa = 3.79GNPAPEE956 pKa = 4.09EE957 pKa = 4.01FTVNGQTCAA966 pKa = 3.44
MM1 pKa = 7.42SPSSSRR7 pKa = 11.84SRR9 pKa = 11.84FAAGAVTAASAIALSMLAGAPGAHH33 pKa = 6.75AAEE36 pKa = 4.69LVTNGGFDD44 pKa = 3.45NGTTGWWNTEE54 pKa = 3.64NAPAEE59 pKa = 4.34VADD62 pKa = 4.29GRR64 pKa = 11.84LCADD68 pKa = 3.72VEE70 pKa = 4.5GGTANAWDD78 pKa = 5.1AIVGQDD84 pKa = 4.84GIALTSGEE92 pKa = 4.4SYY94 pKa = 10.96EE95 pKa = 4.3FTFTASASEE104 pKa = 4.31GTTVRR109 pKa = 11.84ALIQEE114 pKa = 3.99PTEE117 pKa = 4.43PYY119 pKa = 9.73TAFLTANPALTSEE132 pKa = 4.5TQTFSHH138 pKa = 7.02TFTADD143 pKa = 2.85ATLDD147 pKa = 3.63DD148 pKa = 4.44AQLVFQLGGEE158 pKa = 4.14AEE160 pKa = 4.08AWTFCLDD167 pKa = 4.62DD168 pKa = 4.65VSLQDD173 pKa = 3.54GAEE176 pKa = 4.08PPVYY180 pKa = 10.25EE181 pKa = 5.39PDD183 pKa = 2.95TGPPVKK189 pKa = 10.39VNQVGYY195 pKa = 10.69LPDD198 pKa = 3.93GPKK201 pKa = 10.39NATLVTDD208 pKa = 4.1ATSAQPWTLRR218 pKa = 11.84DD219 pKa = 3.23ADD221 pKa = 3.87GAAVAEE227 pKa = 4.93GQTSPQGTEE236 pKa = 3.55QSSGQNVHH244 pKa = 6.84SIDD247 pKa = 3.67FSDD250 pKa = 3.88YY251 pKa = 8.24TEE253 pKa = 4.61KK254 pKa = 11.12GSGYY258 pKa = 8.19TLAVGDD264 pKa = 4.52DD265 pKa = 3.59VSHH268 pKa = 7.21PFDD271 pKa = 3.73ISGSAYY277 pKa = 9.65KK278 pKa = 10.41DD279 pKa = 3.35LRR281 pKa = 11.84TDD283 pKa = 3.43SLSLYY288 pKa = 8.96YY289 pKa = 9.36PQRR292 pKa = 11.84SGIEE296 pKa = 3.81IDD298 pKa = 3.92DD299 pKa = 4.04SLMPGYY305 pKa = 10.28GRR307 pKa = 11.84AAGHH311 pKa = 6.28VGVEE315 pKa = 4.23PNQGDD320 pKa = 3.96LSVPCAPDD328 pKa = 3.18SGCDD332 pKa = 3.53YY333 pKa = 10.94EE334 pKa = 6.2LDD336 pKa = 4.33VSGGWYY342 pKa = 10.02DD343 pKa = 3.88AGDD346 pKa = 3.49HH347 pKa = 6.07GKK349 pKa = 10.25YY350 pKa = 9.61VVNGGISASQVMSVWEE366 pKa = 4.04RR367 pKa = 11.84TTYY370 pKa = 11.23ADD372 pKa = 3.19SASPDD377 pKa = 3.35RR378 pKa = 11.84LGDD381 pKa = 3.3GTLPVPEE388 pKa = 4.72QGNDD392 pKa = 3.28VPDD395 pKa = 3.92VLDD398 pKa = 3.64EE399 pKa = 4.8ARR401 pKa = 11.84WEE403 pKa = 4.06MDD405 pKa = 3.19FLMKK409 pKa = 9.91MQVPEE414 pKa = 4.27GEE416 pKa = 4.34EE417 pKa = 3.79LAGMAHH423 pKa = 6.57HH424 pKa = 6.95KK425 pKa = 9.3MHH427 pKa = 7.0DD428 pKa = 4.03EE429 pKa = 4.01EE430 pKa = 4.82WTGLPLMPADD440 pKa = 4.69DD441 pKa = 4.28PQPRR445 pKa = 11.84YY446 pKa = 9.43LQPPSTAATLNLAATGAQCARR467 pKa = 11.84VFEE470 pKa = 5.01SYY472 pKa = 10.87DD473 pKa = 3.25SEE475 pKa = 4.31FADD478 pKa = 3.59RR479 pKa = 11.84CLEE482 pKa = 4.09TAEE485 pKa = 4.9KK486 pKa = 10.02AWQAAQEE493 pKa = 4.29NPDD496 pKa = 3.68VLADD500 pKa = 3.78PEE502 pKa = 4.31NGVGGGAYY510 pKa = 10.23SDD512 pKa = 5.21DD513 pKa = 3.83DD514 pKa = 4.72VSDD517 pKa = 3.53EE518 pKa = 4.94FYY520 pKa = 10.34WAAAEE525 pKa = 4.28LFITTGEE532 pKa = 3.91QSYY535 pKa = 11.18EE536 pKa = 4.06DD537 pKa = 3.6AVLDD541 pKa = 4.34HH542 pKa = 6.89EE543 pKa = 4.93LHH545 pKa = 6.05TEE547 pKa = 4.07DD548 pKa = 4.55VFTAGGLAWGAVAPLGRR565 pKa = 11.84MNLALVPNKK574 pKa = 10.33LDD576 pKa = 3.68DD577 pKa = 3.94RR578 pKa = 11.84DD579 pKa = 3.78RR580 pKa = 11.84VVDD583 pKa = 4.07SVVAGADD590 pKa = 3.51EE591 pKa = 4.16YY592 pKa = 11.71LDD594 pKa = 3.53TLRR597 pKa = 11.84NHH599 pKa = 7.2AFGLPYY605 pKa = 10.52APPDD609 pKa = 3.52NVFVWGSNSQVLNNMVVMTAAYY631 pKa = 10.33DD632 pKa = 3.72LTGDD636 pKa = 3.57GAYY639 pKa = 10.07RR640 pKa = 11.84DD641 pKa = 4.1GVLEE645 pKa = 4.21GMDD648 pKa = 3.88YY649 pKa = 10.92ILGRR653 pKa = 11.84NALGQSYY660 pKa = 7.56VTGYY664 pKa = 11.12GEE666 pKa = 3.97QFSKK670 pKa = 10.56NQHH673 pKa = 4.93SRR675 pKa = 11.84WYY677 pKa = 10.58ANQLDD682 pKa = 4.36ADD684 pKa = 4.48LPNPPEE690 pKa = 4.32GTVSGGPNSQTSTWDD705 pKa = 3.09PVAQDD710 pKa = 4.0KK711 pKa = 11.73LEE713 pKa = 4.31GCAPQFCYY721 pKa = 10.09IDD723 pKa = 5.73DD724 pKa = 4.03IEE726 pKa = 4.27SWATNEE732 pKa = 5.0LTVNWNAPLAIVSSFIADD750 pKa = 3.36QTGEE754 pKa = 4.23GDD756 pKa = 3.84PPAEE760 pKa = 5.1DD761 pKa = 3.48EE762 pKa = 4.53TAPTAPGKK770 pKa = 9.16PQVSGVTSSQAEE782 pKa = 4.45LTWEE786 pKa = 4.06ASTDD790 pKa = 3.44EE791 pKa = 4.73GGSGVEE797 pKa = 4.61GYY799 pKa = 10.24DD800 pKa = 3.44VYY802 pKa = 11.57AGTGDD807 pKa = 3.75GAQKK811 pKa = 10.49VGSASGPSFTLTGLEE826 pKa = 4.28SEE828 pKa = 4.67TEE830 pKa = 4.24YY831 pKa = 10.5TVHH834 pKa = 6.25VVARR838 pKa = 11.84DD839 pKa = 3.52GAGNRR844 pKa = 11.84SEE846 pKa = 4.81PGPNAAFTTEE856 pKa = 3.69AGGTGGEE863 pKa = 4.24AACEE867 pKa = 3.82VDD869 pKa = 4.71YY870 pKa = 10.42STNDD874 pKa = 2.96WNGGFTASVSVVNTGDD890 pKa = 3.29TPIRR894 pKa = 11.84DD895 pKa = 3.38WEE897 pKa = 4.37LGFEE901 pKa = 4.7FGAGQEE907 pKa = 4.24ITHH910 pKa = 6.43GWNAEE915 pKa = 3.61WSQDD919 pKa = 3.14GSAVTASGLSWNSDD933 pKa = 3.7LAPGSSVSAGFNGTAQDD950 pKa = 3.79GNPAPEE956 pKa = 4.09EE957 pKa = 4.01FTVNGQTCAA966 pKa = 3.44
Molecular weight: 101.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C2FMM6|A0A0C2FMM6_9ACTN Hydrolase OS=Streptomonospora alba OX=183763 GN=LP52_00515 PE=3 SV=1
MM1 pKa = 7.49RR2 pKa = 11.84HH3 pKa = 5.9SLLGGDD9 pKa = 3.6RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84PLGFGRR19 pKa = 11.84LTRR22 pKa = 11.84IGAVAGGAVARR33 pKa = 11.84GNIAVGVAVRR43 pKa = 11.84GTVRR47 pKa = 11.84TVARR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84VHH55 pKa = 6.08IPVRR59 pKa = 11.84RR60 pKa = 11.84GVRR63 pKa = 11.84IPVRR67 pKa = 11.84GAEE70 pKa = 4.04PVAVVAARR78 pKa = 11.84VSAVSVAAGTAAGVVRR94 pKa = 11.84LL95 pKa = 4.12
MM1 pKa = 7.49RR2 pKa = 11.84HH3 pKa = 5.9SLLGGDD9 pKa = 3.6RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84PLGFGRR19 pKa = 11.84LTRR22 pKa = 11.84IGAVAGGAVARR33 pKa = 11.84GNIAVGVAVRR43 pKa = 11.84GTVRR47 pKa = 11.84TVARR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84VHH55 pKa = 6.08IPVRR59 pKa = 11.84RR60 pKa = 11.84GVRR63 pKa = 11.84IPVRR67 pKa = 11.84GAEE70 pKa = 4.04PVAVVAARR78 pKa = 11.84VSAVSVAAGTAAGVVRR94 pKa = 11.84LL95 pKa = 4.12
Molecular weight: 9.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1345555 |
28 |
2499 |
312.4 |
33.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.665 ± 0.062 | 0.774 ± 0.011 |
6.261 ± 0.036 | 6.575 ± 0.036 |
2.78 ± 0.021 | 9.574 ± 0.04 |
2.198 ± 0.017 | 3.281 ± 0.024 |
1.599 ± 0.022 | 10.053 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.946 ± 0.015 | 1.727 ± 0.018 |
5.834 ± 0.033 | 2.695 ± 0.024 |
8.271 ± 0.038 | 5.317 ± 0.029 |
5.468 ± 0.024 | 8.492 ± 0.029 |
1.428 ± 0.015 | 2.063 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
