
Marinobacter mobilis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Marinobacter
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
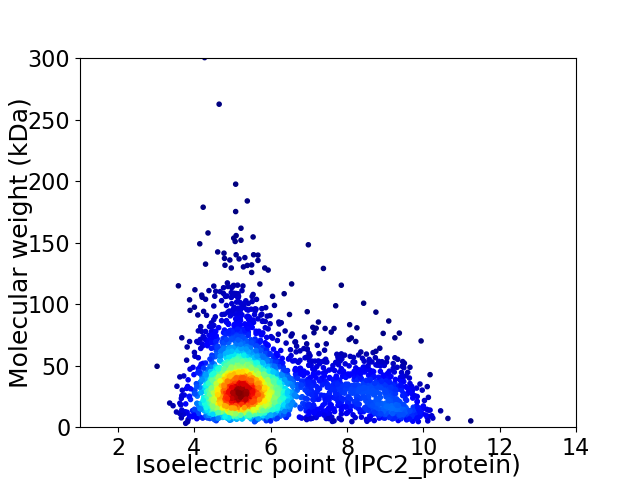
Virtual 2D-PAGE plot for 3442 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3DCN1|A0A1H3DCN1_9ALTE DNA-binding transcriptional response regulator NtrC family contains REC AAA-type ATPase and a Fis-type DNA-binding domains OS=Marinobacter mobilis OX=488533 GN=SAMN04487960_11226 PE=4 SV=1
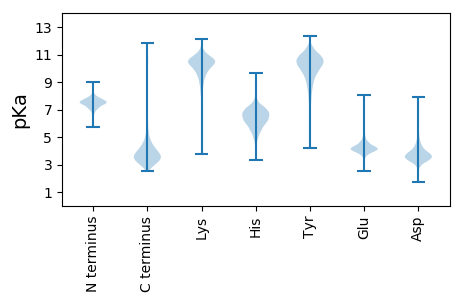
MM1 pKa = 7.09QNSAALWTLPLALTLAACNVDD22 pKa = 2.82ISTTEE27 pKa = 3.88VTPGQPDD34 pKa = 3.81SEE36 pKa = 4.76TPTPVKK42 pKa = 10.71DD43 pKa = 3.67LDD45 pKa = 3.86SDD47 pKa = 4.32GVADD51 pKa = 4.77SADD54 pKa = 3.44NCLLVPNADD63 pKa = 3.47QANSNGDD70 pKa = 3.87ALGDD74 pKa = 3.71ACDD77 pKa = 3.47IDD79 pKa = 4.2TPVVEE84 pKa = 5.1GKK86 pKa = 9.91YY87 pKa = 10.19QLTIYY92 pKa = 10.38HH93 pKa = 6.62EE94 pKa = 4.74LGSEE98 pKa = 4.16YY99 pKa = 10.78LDD101 pKa = 3.87AASGVCVSATDD112 pKa = 3.49SPFTVDD118 pKa = 3.53VLVKK122 pKa = 10.24GSQVFLSEE130 pKa = 4.06EE131 pKa = 3.83GGEE134 pKa = 3.9RR135 pKa = 11.84RR136 pKa = 11.84FYY138 pKa = 11.26GIADD142 pKa = 3.4TSGNLEE148 pKa = 4.34VFTHH152 pKa = 6.89PEE154 pKa = 3.79GQFAATNGYY163 pKa = 9.43FSEE166 pKa = 4.44GTRR169 pKa = 11.84RR170 pKa = 11.84ITFDD174 pKa = 3.98YY175 pKa = 10.97SDD177 pKa = 3.51QQSNLDD183 pKa = 3.61GTVSCNIAARR193 pKa = 11.84VEE195 pKa = 4.55ANFNYY200 pKa = 8.24ATSLIAPAAMVPPGLSPGDD219 pKa = 3.22TFYY222 pKa = 10.94IVFVTSAGTVANLSVDD238 pKa = 3.38AFNLFVNDD246 pKa = 3.53VADD249 pKa = 4.42GSSLKK254 pKa = 10.14GTDD257 pKa = 3.55NGDD260 pKa = 4.01LRR262 pKa = 11.84WKK264 pKa = 10.54AILGHH269 pKa = 7.42DD270 pKa = 4.4DD271 pKa = 3.4GTIQGEE277 pKa = 4.24NLFSVGSTSPIYY289 pKa = 10.47NLNGDD294 pKa = 3.91RR295 pKa = 11.84LANNASDD302 pKa = 4.09FFNGGMLNPIEE313 pKa = 4.15YY314 pKa = 10.26DD315 pKa = 3.31EE316 pKa = 5.09TGTSLGDD323 pKa = 3.43VNVSVGLTQTGSNLLRR339 pKa = 11.84QGDD342 pKa = 3.84NTLGGNDD349 pKa = 3.88FMGDD353 pKa = 2.94GCSRR357 pKa = 11.84GRR359 pKa = 11.84SGQINKK365 pKa = 9.32PAFAASISGDD375 pKa = 4.0DD376 pKa = 4.14FCDD379 pKa = 3.34STPYY383 pKa = 10.27PLYY386 pKa = 10.59SVSPLLEE393 pKa = 4.24VPSAPP398 pKa = 4.25
MM1 pKa = 7.09QNSAALWTLPLALTLAACNVDD22 pKa = 2.82ISTTEE27 pKa = 3.88VTPGQPDD34 pKa = 3.81SEE36 pKa = 4.76TPTPVKK42 pKa = 10.71DD43 pKa = 3.67LDD45 pKa = 3.86SDD47 pKa = 4.32GVADD51 pKa = 4.77SADD54 pKa = 3.44NCLLVPNADD63 pKa = 3.47QANSNGDD70 pKa = 3.87ALGDD74 pKa = 3.71ACDD77 pKa = 3.47IDD79 pKa = 4.2TPVVEE84 pKa = 5.1GKK86 pKa = 9.91YY87 pKa = 10.19QLTIYY92 pKa = 10.38HH93 pKa = 6.62EE94 pKa = 4.74LGSEE98 pKa = 4.16YY99 pKa = 10.78LDD101 pKa = 3.87AASGVCVSATDD112 pKa = 3.49SPFTVDD118 pKa = 3.53VLVKK122 pKa = 10.24GSQVFLSEE130 pKa = 4.06EE131 pKa = 3.83GGEE134 pKa = 3.9RR135 pKa = 11.84RR136 pKa = 11.84FYY138 pKa = 11.26GIADD142 pKa = 3.4TSGNLEE148 pKa = 4.34VFTHH152 pKa = 6.89PEE154 pKa = 3.79GQFAATNGYY163 pKa = 9.43FSEE166 pKa = 4.44GTRR169 pKa = 11.84RR170 pKa = 11.84ITFDD174 pKa = 3.98YY175 pKa = 10.97SDD177 pKa = 3.51QQSNLDD183 pKa = 3.61GTVSCNIAARR193 pKa = 11.84VEE195 pKa = 4.55ANFNYY200 pKa = 8.24ATSLIAPAAMVPPGLSPGDD219 pKa = 3.22TFYY222 pKa = 10.94IVFVTSAGTVANLSVDD238 pKa = 3.38AFNLFVNDD246 pKa = 3.53VADD249 pKa = 4.42GSSLKK254 pKa = 10.14GTDD257 pKa = 3.55NGDD260 pKa = 4.01LRR262 pKa = 11.84WKK264 pKa = 10.54AILGHH269 pKa = 7.42DD270 pKa = 4.4DD271 pKa = 3.4GTIQGEE277 pKa = 4.24NLFSVGSTSPIYY289 pKa = 10.47NLNGDD294 pKa = 3.91RR295 pKa = 11.84LANNASDD302 pKa = 4.09FFNGGMLNPIEE313 pKa = 4.15YY314 pKa = 10.26DD315 pKa = 3.31EE316 pKa = 5.09TGTSLGDD323 pKa = 3.43VNVSVGLTQTGSNLLRR339 pKa = 11.84QGDD342 pKa = 3.84NTLGGNDD349 pKa = 3.88FMGDD353 pKa = 2.94GCSRR357 pKa = 11.84GRR359 pKa = 11.84SGQINKK365 pKa = 9.32PAFAASISGDD375 pKa = 4.0DD376 pKa = 4.14FCDD379 pKa = 3.34STPYY383 pKa = 10.27PLYY386 pKa = 10.59SVSPLLEE393 pKa = 4.24VPSAPP398 pKa = 4.25
Molecular weight: 41.67 kDa
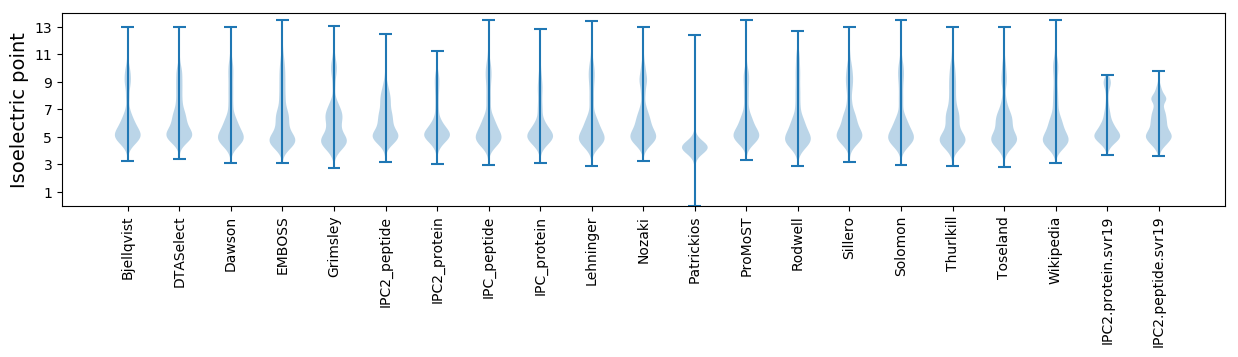
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H2WHE6|A0A1H2WHE6_9ALTE Amino acid ABC transporter substrate-binding protein PAAT family (TC 3.A.1.3.-) OS=Marinobacter mobilis OX=488533 GN=SAMN04487960_104170 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.88ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.4GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.88ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.4GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1144372 |
29 |
2741 |
332.5 |
36.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.075 ± 0.046 | 0.975 ± 0.012 |
5.893 ± 0.027 | 6.272 ± 0.037 |
3.678 ± 0.028 | 7.849 ± 0.035 |
2.269 ± 0.022 | 5.071 ± 0.029 |
3.152 ± 0.028 | 10.945 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.489 ± 0.018 | 3.23 ± 0.024 |
4.795 ± 0.031 | 4.3 ± 0.031 |
6.609 ± 0.037 | 5.991 ± 0.025 |
5.119 ± 0.029 | 7.309 ± 0.034 |
1.369 ± 0.016 | 2.611 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
