
Dyella sp. AD56
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Rhodanobacteraceae; Dyella; unclassified Dyella
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
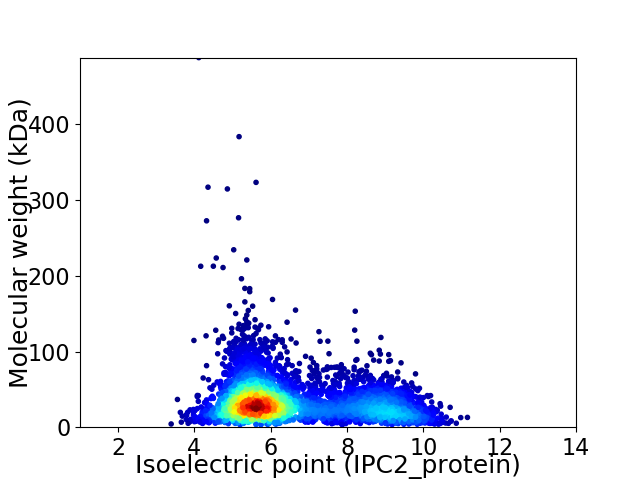
Virtual 2D-PAGE plot for 4642 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N7QY61|A0A2N7QY61_9GAMM DNA polymerase IV OS=Dyella sp. AD56 OX=1528744 GN=dinB_1 PE=3 SV=1
MM1 pKa = 6.59NTIYY5 pKa = 11.08SKK7 pKa = 10.21IWNSALGAVVVASEE21 pKa = 4.45LARR24 pKa = 11.84MAGKK28 pKa = 10.17GRR30 pKa = 11.84QRR32 pKa = 11.84RR33 pKa = 11.84ARR35 pKa = 11.84TLTVIGVLVLSMAMANGVQAGVTLPIATPPTTTPASGTTTTVVDD79 pKa = 3.53PTGAVAISGPASGGTTTASTAGSNDD104 pKa = 3.13VAVGSGAVTLGSNGTAVGAGAQSQGDD130 pKa = 3.59NGTSLGAAANATATGANSTAVGTGAQSQGDD160 pKa = 3.58NGTAVGSAANAVGNAAVAVGANATATGMNSVALGAGAAAATDD202 pKa = 3.4SSMALGQGAQATGSAAMAMGAAAGATGSNSMALGQGAQATASNSIALGNSTVAASSSSIAMGSSASVTGVSSSVAIGPGASITNQTLAGTTAFGFYY298 pKa = 10.28RR299 pKa = 11.84NDD301 pKa = 2.85VAMGNGASLYY311 pKa = 10.62AATSSVAIGAQSFVGDD327 pKa = 4.1TSATPALVTSATAVGVGSSVQASYY351 pKa = 8.64GTAIGDD357 pKa = 3.62IASTSGTYY365 pKa = 8.42ATAVGYY371 pKa = 9.29GASSVGNTASAFGSGASAFGAGSTALGGNAQANGAGSIVIAGGGNGGGAYY421 pKa = 10.19SGPNSPNTVVIGNSAEE437 pKa = 4.13AVEE440 pKa = 4.66TTDD443 pKa = 4.49SNGNAQLVSNSVVIGSNAISYY464 pKa = 10.61GGLNGIAIGNTAGVAQGSIDD484 pKa = 4.15AIGIGTRR491 pKa = 11.84AIGYY495 pKa = 9.76GEE497 pKa = 4.54GSVAMGAGSYY507 pKa = 10.65AIGPGSTAVGGWLDD521 pKa = 3.4INGTGVVTGAGSGTTVSPGVGSSAFGAGAEE551 pKa = 4.43SIGDD555 pKa = 3.38FSVAMGAGATAAGSSTGYY573 pKa = 10.0PNANRR578 pKa = 11.84AGYY581 pKa = 8.5TNADD585 pKa = 3.37QTAGAIAIGYY595 pKa = 8.15EE596 pKa = 4.44SYY598 pKa = 11.27TNGDD602 pKa = 3.27GSVVIGRR609 pKa = 11.84NALANGSPDD618 pKa = 2.91GSLINGTAIGSQSVAWGTGAVAIGSSAVAAWTNDD652 pKa = 2.81QAMGTNAYY660 pKa = 10.09AGGGTGTNNMAIGNGATAGDD680 pKa = 4.02ALRR683 pKa = 11.84GTEE686 pKa = 4.07NNQNPPLPLPNISDD700 pKa = 3.72TVAIGASSSATANQAMALGVSASAAGVQAQAFGNNAIANGVQSMSNGYY748 pKa = 8.17MADD751 pKa = 3.54AEE753 pKa = 4.79GDD755 pKa = 3.48YY756 pKa = 11.76SMAQGSGAYY765 pKa = 9.62AAGTGDD771 pKa = 3.35VALGSGAVATGANGMGTAVGYY792 pKa = 9.63GATVTAQNAVAIGANSVADD811 pKa = 3.97RR812 pKa = 11.84DD813 pKa = 3.97NAVSVGAAGAEE824 pKa = 3.65RR825 pKa = 11.84QIINVAAGTQATDD838 pKa = 3.45AVNLSQLQAAGLVDD852 pKa = 4.19GNNNPLTAVTYY863 pKa = 11.11DD864 pKa = 3.6DD865 pKa = 3.71ATKK868 pKa = 10.86AGITLQGANGTQIHH882 pKa = 5.83NVAAGVAPMDD892 pKa = 4.43AVNMGQFTSATSTINGEE909 pKa = 4.16LSTLTTDD916 pKa = 4.69FTTLTQQLGAGGGGMPGTFAVNGQGGSMTNASANGANTVAIGNGAAVGTGVSGDD970 pKa = 3.16NGTAVGGGALAHH982 pKa = 6.57GPNDD986 pKa = 3.53TAFGGNAKK994 pKa = 10.0VNADD998 pKa = 2.9GSTAVGANTTIAVAATNAVAVGEE1021 pKa = 4.53SASVTAASGTAVGQGSSATATNTVALGQGSVANQANTVSVGTATNQRR1068 pKa = 11.84QIVNVAAGVNATDD1081 pKa = 3.66ATNVGQVSAQVTQALQTAKK1100 pKa = 9.77TYY1102 pKa = 10.68TDD1104 pKa = 3.47ASSQQAVQSANAYY1117 pKa = 6.89TNQAIANYY1125 pKa = 9.0DD1126 pKa = 3.61ANSGIGALRR1135 pKa = 11.84QEE1137 pKa = 4.9MNDD1140 pKa = 3.0QFNNVNQRR1148 pKa = 11.84LDD1150 pKa = 3.2RR1151 pKa = 11.84VGAMGSAMAQMTANTSNLAGDD1172 pKa = 3.56NRR1174 pKa = 11.84VGFGAGNYY1182 pKa = 8.99NGQGAFAVGYY1192 pKa = 9.39QRR1194 pKa = 11.84AFASSHH1200 pKa = 5.84ASVSVGASVSGSEE1213 pKa = 4.12TSVGVGGGFSWW1224 pKa = 3.94
MM1 pKa = 6.59NTIYY5 pKa = 11.08SKK7 pKa = 10.21IWNSALGAVVVASEE21 pKa = 4.45LARR24 pKa = 11.84MAGKK28 pKa = 10.17GRR30 pKa = 11.84QRR32 pKa = 11.84RR33 pKa = 11.84ARR35 pKa = 11.84TLTVIGVLVLSMAMANGVQAGVTLPIATPPTTTPASGTTTTVVDD79 pKa = 3.53PTGAVAISGPASGGTTTASTAGSNDD104 pKa = 3.13VAVGSGAVTLGSNGTAVGAGAQSQGDD130 pKa = 3.59NGTSLGAAANATATGANSTAVGTGAQSQGDD160 pKa = 3.58NGTAVGSAANAVGNAAVAVGANATATGMNSVALGAGAAAATDD202 pKa = 3.4SSMALGQGAQATGSAAMAMGAAAGATGSNSMALGQGAQATASNSIALGNSTVAASSSSIAMGSSASVTGVSSSVAIGPGASITNQTLAGTTAFGFYY298 pKa = 10.28RR299 pKa = 11.84NDD301 pKa = 2.85VAMGNGASLYY311 pKa = 10.62AATSSVAIGAQSFVGDD327 pKa = 4.1TSATPALVTSATAVGVGSSVQASYY351 pKa = 8.64GTAIGDD357 pKa = 3.62IASTSGTYY365 pKa = 8.42ATAVGYY371 pKa = 9.29GASSVGNTASAFGSGASAFGAGSTALGGNAQANGAGSIVIAGGGNGGGAYY421 pKa = 10.19SGPNSPNTVVIGNSAEE437 pKa = 4.13AVEE440 pKa = 4.66TTDD443 pKa = 4.49SNGNAQLVSNSVVIGSNAISYY464 pKa = 10.61GGLNGIAIGNTAGVAQGSIDD484 pKa = 4.15AIGIGTRR491 pKa = 11.84AIGYY495 pKa = 9.76GEE497 pKa = 4.54GSVAMGAGSYY507 pKa = 10.65AIGPGSTAVGGWLDD521 pKa = 3.4INGTGVVTGAGSGTTVSPGVGSSAFGAGAEE551 pKa = 4.43SIGDD555 pKa = 3.38FSVAMGAGATAAGSSTGYY573 pKa = 10.0PNANRR578 pKa = 11.84AGYY581 pKa = 8.5TNADD585 pKa = 3.37QTAGAIAIGYY595 pKa = 8.15EE596 pKa = 4.44SYY598 pKa = 11.27TNGDD602 pKa = 3.27GSVVIGRR609 pKa = 11.84NALANGSPDD618 pKa = 2.91GSLINGTAIGSQSVAWGTGAVAIGSSAVAAWTNDD652 pKa = 2.81QAMGTNAYY660 pKa = 10.09AGGGTGTNNMAIGNGATAGDD680 pKa = 4.02ALRR683 pKa = 11.84GTEE686 pKa = 4.07NNQNPPLPLPNISDD700 pKa = 3.72TVAIGASSSATANQAMALGVSASAAGVQAQAFGNNAIANGVQSMSNGYY748 pKa = 8.17MADD751 pKa = 3.54AEE753 pKa = 4.79GDD755 pKa = 3.48YY756 pKa = 11.76SMAQGSGAYY765 pKa = 9.62AAGTGDD771 pKa = 3.35VALGSGAVATGANGMGTAVGYY792 pKa = 9.63GATVTAQNAVAIGANSVADD811 pKa = 3.97RR812 pKa = 11.84DD813 pKa = 3.97NAVSVGAAGAEE824 pKa = 3.65RR825 pKa = 11.84QIINVAAGTQATDD838 pKa = 3.45AVNLSQLQAAGLVDD852 pKa = 4.19GNNNPLTAVTYY863 pKa = 11.11DD864 pKa = 3.6DD865 pKa = 3.71ATKK868 pKa = 10.86AGITLQGANGTQIHH882 pKa = 5.83NVAAGVAPMDD892 pKa = 4.43AVNMGQFTSATSTINGEE909 pKa = 4.16LSTLTTDD916 pKa = 4.69FTTLTQQLGAGGGGMPGTFAVNGQGGSMTNASANGANTVAIGNGAAVGTGVSGDD970 pKa = 3.16NGTAVGGGALAHH982 pKa = 6.57GPNDD986 pKa = 3.53TAFGGNAKK994 pKa = 10.0VNADD998 pKa = 2.9GSTAVGANTTIAVAATNAVAVGEE1021 pKa = 4.53SASVTAASGTAVGQGSSATATNTVALGQGSVANQANTVSVGTATNQRR1068 pKa = 11.84QIVNVAAGVNATDD1081 pKa = 3.66ATNVGQVSAQVTQALQTAKK1100 pKa = 9.77TYY1102 pKa = 10.68TDD1104 pKa = 3.47ASSQQAVQSANAYY1117 pKa = 6.89TNQAIANYY1125 pKa = 9.0DD1126 pKa = 3.61ANSGIGALRR1135 pKa = 11.84QEE1137 pKa = 4.9MNDD1140 pKa = 3.0QFNNVNQRR1148 pKa = 11.84LDD1150 pKa = 3.2RR1151 pKa = 11.84VGAMGSAMAQMTANTSNLAGDD1172 pKa = 3.56NRR1174 pKa = 11.84VGFGAGNYY1182 pKa = 8.99NGQGAFAVGYY1192 pKa = 9.39QRR1194 pKa = 11.84AFASSHH1200 pKa = 5.84ASVSVGASVSGSEE1213 pKa = 4.12TSVGVGGGFSWW1224 pKa = 3.94
Molecular weight: 114.63 kDa
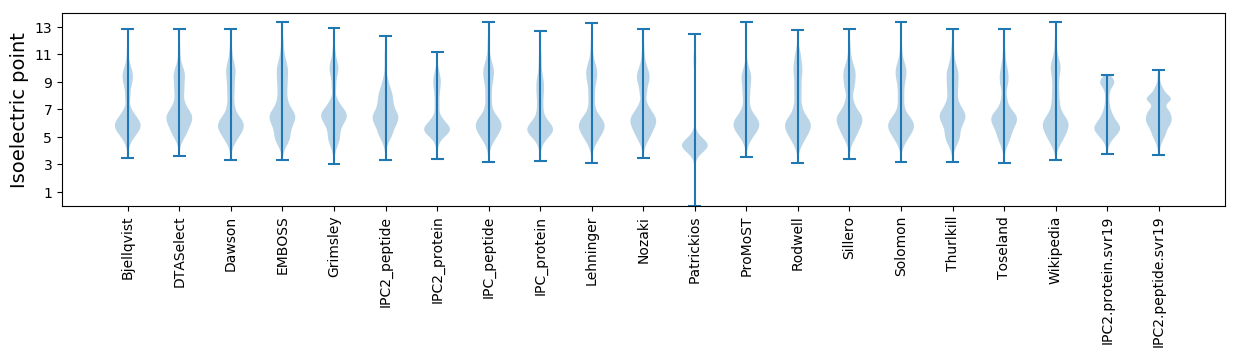
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N7QVH0|A0A2N7QVH0_9GAMM 2-dehydropantoate 2-reductase OS=Dyella sp. AD56 OX=1528744 GN=panE PE=3 SV=1
MM1 pKa = 7.73AKK3 pKa = 6.84TTRR6 pKa = 11.84SATATKK12 pKa = 8.9PQKK15 pKa = 10.45GKK17 pKa = 9.35PAGKK21 pKa = 10.41VKK23 pKa = 10.43DD24 pKa = 3.74VKK26 pKa = 10.74AGKK29 pKa = 9.65KK30 pKa = 8.62VAAGRR35 pKa = 11.84SAVPTKK41 pKa = 10.45AAAVKK46 pKa = 10.4KK47 pKa = 9.45PAVKK51 pKa = 10.21NAPVKK56 pKa = 10.34KK57 pKa = 10.04AAAAKK62 pKa = 9.82KK63 pKa = 8.4PAAAKK68 pKa = 10.18VVAKK72 pKa = 10.76APAKK76 pKa = 10.38SVAKK80 pKa = 10.08KK81 pKa = 9.98AAPKK85 pKa = 10.18PPAKK89 pKa = 10.04KK90 pKa = 10.02ARR92 pKa = 11.84RR93 pKa = 11.84PRR95 pKa = 11.84NRR97 pKa = 11.84PSLRR101 pKa = 11.84KK102 pKa = 9.57LRR104 pKa = 11.84NRR106 pKa = 11.84SPRR109 pKa = 11.84KK110 pKa = 9.03FRR112 pKa = 11.84SPPRR116 pKa = 11.84RR117 pKa = 11.84SRR119 pKa = 11.84PRR121 pKa = 11.84RR122 pKa = 3.42
MM1 pKa = 7.73AKK3 pKa = 6.84TTRR6 pKa = 11.84SATATKK12 pKa = 8.9PQKK15 pKa = 10.45GKK17 pKa = 9.35PAGKK21 pKa = 10.41VKK23 pKa = 10.43DD24 pKa = 3.74VKK26 pKa = 10.74AGKK29 pKa = 9.65KK30 pKa = 8.62VAAGRR35 pKa = 11.84SAVPTKK41 pKa = 10.45AAAVKK46 pKa = 10.4KK47 pKa = 9.45PAVKK51 pKa = 10.21NAPVKK56 pKa = 10.34KK57 pKa = 10.04AAAAKK62 pKa = 9.82KK63 pKa = 8.4PAAAKK68 pKa = 10.18VVAKK72 pKa = 10.76APAKK76 pKa = 10.38SVAKK80 pKa = 10.08KK81 pKa = 9.98AAPKK85 pKa = 10.18PPAKK89 pKa = 10.04KK90 pKa = 10.02ARR92 pKa = 11.84RR93 pKa = 11.84PRR95 pKa = 11.84NRR97 pKa = 11.84PSLRR101 pKa = 11.84KK102 pKa = 9.57LRR104 pKa = 11.84NRR106 pKa = 11.84SPRR109 pKa = 11.84KK110 pKa = 9.03FRR112 pKa = 11.84SPPRR116 pKa = 11.84RR117 pKa = 11.84SRR119 pKa = 11.84PRR121 pKa = 11.84RR122 pKa = 3.42
Molecular weight: 12.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1539333 |
30 |
4705 |
331.6 |
36.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.882 ± 0.045 | 0.831 ± 0.011 |
5.773 ± 0.03 | 4.917 ± 0.042 |
3.423 ± 0.023 | 8.42 ± 0.067 |
2.497 ± 0.018 | 4.365 ± 0.024 |
3.06 ± 0.032 | 10.471 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.365 ± 0.019 | 2.953 ± 0.039 |
5.171 ± 0.028 | 3.928 ± 0.028 |
6.772 ± 0.05 | 5.928 ± 0.034 |
5.521 ± 0.05 | 7.551 ± 0.031 |
1.57 ± 0.02 | 2.601 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
