
Agromyces sp. KACC 19306
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Agromyces
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
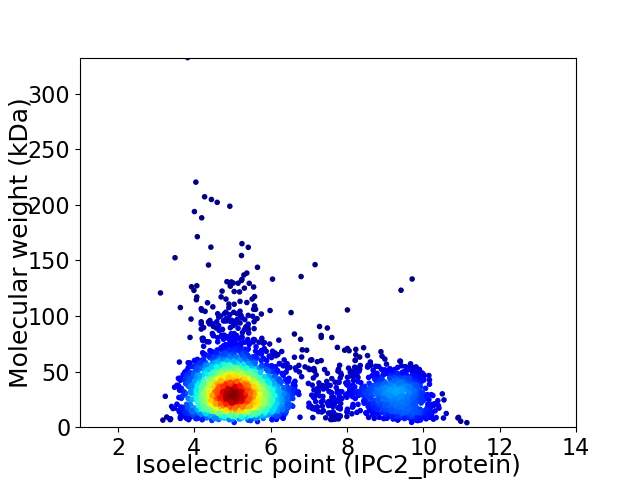
Virtual 2D-PAGE plot for 3349 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
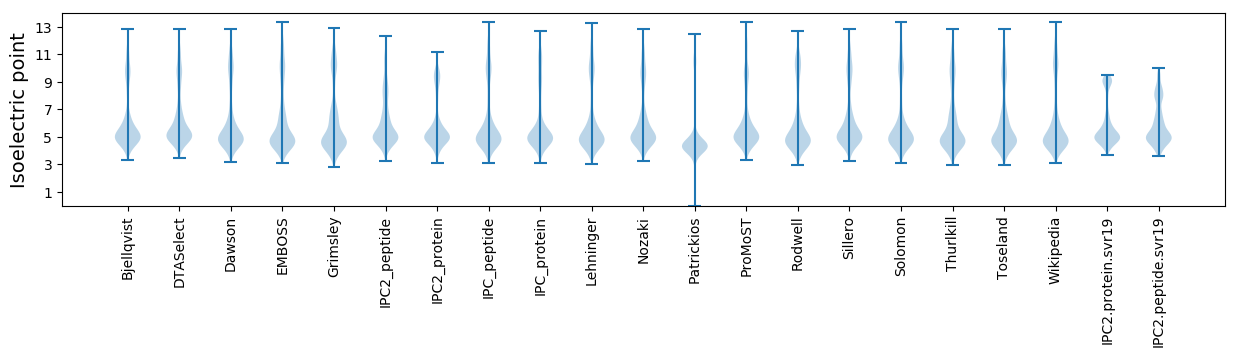
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C1YC09|A0A5C1YC09_9MICO 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase OS=Agromyces sp. KACC 19306 OX=2592652 GN=ispD PE=3 SV=1
MM1 pKa = 7.78ANRR4 pKa = 11.84SAEE7 pKa = 3.95ASIAPPQQPLSCEE20 pKa = 3.99SLKK23 pKa = 10.85PSSEE27 pKa = 4.21LEE29 pKa = 3.98DD30 pKa = 3.06RR31 pKa = 11.84TAYY34 pKa = 10.63LDD36 pKa = 4.0CLASQNAGLRR46 pKa = 11.84SFAAAAAVDD55 pKa = 4.19EE56 pKa = 4.96NGNPDD61 pKa = 4.18LPDD64 pKa = 3.43SCGIDD69 pKa = 2.84IALILDD75 pKa = 3.52KK76 pKa = 11.11SGSIGGAGIANLKK89 pKa = 9.57SAANAFTGALVDD101 pKa = 4.53LGSQVSVVSFDD112 pKa = 3.06QDD114 pKa = 3.21AYY116 pKa = 11.58LLLPATDD123 pKa = 3.79LTSGNLGTIQGSYY136 pKa = 10.59AGLVSDD142 pKa = 5.52GWTNWKK148 pKa = 10.23RR149 pKa = 11.84GLEE152 pKa = 4.1VGLGTFGGFDD162 pKa = 3.56DD163 pKa = 4.37TVEE166 pKa = 4.1LTVVITDD173 pKa = 3.97GNPNTVNPANGGQFPDD189 pKa = 4.56GSPGALDD196 pKa = 3.64PAVLEE201 pKa = 4.34ANAIKK206 pKa = 9.11GTPSHH211 pKa = 6.03VFVVGVGPSVSTGPIVAISGAEE233 pKa = 3.82VLAADD238 pKa = 4.81GSNINTADD246 pKa = 3.48YY247 pKa = 7.46MTTTSYY253 pKa = 10.17ATLAEE258 pKa = 4.41DD259 pKa = 3.79LRR261 pKa = 11.84AVATALCGGTVVVHH275 pKa = 6.35KK276 pKa = 10.81SVDD279 pKa = 3.57GVATGGWGFEE289 pKa = 4.35TPTTDD294 pKa = 3.09VTPAALTTLADD305 pKa = 3.4GTGIFEE311 pKa = 4.36VEE313 pKa = 4.62GYY315 pKa = 9.88DD316 pKa = 3.25AQTDD320 pKa = 3.7VRR322 pKa = 11.84TVTITEE328 pKa = 4.03EE329 pKa = 3.83TRR331 pKa = 11.84PNHH334 pKa = 5.61GLEE337 pKa = 4.06NLYY340 pKa = 10.96CEE342 pKa = 4.97LNGEE346 pKa = 4.35QVPITHH352 pKa = 7.64DD353 pKa = 3.45EE354 pKa = 4.42GSPTWSVDD362 pKa = 3.14VGKK365 pKa = 10.76LDD367 pKa = 4.9IVHH370 pKa = 6.47CWVEE374 pKa = 3.9NEE376 pKa = 4.01TAKK379 pKa = 10.63WEE381 pKa = 4.22VTKK384 pKa = 10.73TSDD387 pKa = 3.86PPSGTAVEE395 pKa = 4.51AGDD398 pKa = 4.2FIDD401 pKa = 3.77YY402 pKa = 8.59TLHH405 pKa = 6.09IRR407 pKa = 11.84HH408 pKa = 6.4LSGPNVTDD416 pKa = 4.33FDD418 pKa = 4.37IDD420 pKa = 4.29DD421 pKa = 6.01DD422 pKa = 4.23ISQLAPYY429 pKa = 10.17VSFVGLLTPAPVVSSDD445 pKa = 2.76WDD447 pKa = 3.76GTPGKK452 pKa = 10.3YY453 pKa = 9.94LLKK456 pKa = 10.48LAQLTKK462 pKa = 10.91AEE464 pKa = 4.32TLNIRR469 pKa = 11.84YY470 pKa = 8.81RR471 pKa = 11.84VQVDD475 pKa = 3.02ADD477 pKa = 3.91VPPGSVLRR485 pKa = 11.84NVVLTNCPAPAEE497 pKa = 4.46GEE499 pKa = 3.83PDD501 pKa = 3.3LCVTEE506 pKa = 4.62NPTPGWEE513 pKa = 3.83LRR515 pKa = 11.84KK516 pKa = 10.17YY517 pKa = 10.52SNPPSGSVVYY527 pKa = 9.78PGYY530 pKa = 10.59EE531 pKa = 3.67VTYY534 pKa = 10.51EE535 pKa = 3.58LDD537 pKa = 3.31AYY539 pKa = 10.3NFSEE543 pKa = 4.48APVIGGIATDD553 pKa = 4.37DD554 pKa = 4.11LSDD557 pKa = 3.7VLDD560 pKa = 3.88NATIDD565 pKa = 3.63EE566 pKa = 5.07LGPGLTLEE574 pKa = 4.54GTTLTWAVPDD584 pKa = 4.67LEE586 pKa = 6.13PGDD589 pKa = 4.43SVSTSYY595 pKa = 10.46TVIVNDD601 pKa = 4.49DD602 pKa = 2.94AWGQTLTNVVTPEE615 pKa = 4.19YY616 pKa = 9.83PGSCPPPPQDD626 pKa = 3.34GEE628 pKa = 4.7VVVLAEE634 pKa = 4.59GEE636 pKa = 4.48CTTDD640 pKa = 5.03HH641 pKa = 6.61EE642 pKa = 4.81VADD645 pKa = 4.27PNLRR649 pKa = 11.84IQKK652 pKa = 9.61LWEE655 pKa = 4.38HH656 pKa = 7.23DD657 pKa = 3.76GGDD660 pKa = 3.73NPVDD664 pKa = 4.18SGDD667 pKa = 4.53DD668 pKa = 3.62PPDD671 pKa = 3.86VISYY675 pKa = 10.72AIGVWNSGNAPVEE688 pKa = 4.31NPVVTDD694 pKa = 3.65TLPEE698 pKa = 4.2GLTFVPGSEE707 pKa = 4.19VVPAGWVLDD716 pKa = 3.8ASVPGQVTFSQEE728 pKa = 3.85TPGPFDD734 pKa = 5.11PIAYY738 pKa = 8.49PGVIFEE744 pKa = 4.72FDD746 pKa = 3.73VEE748 pKa = 4.45VGEE751 pKa = 5.0LAQPDD756 pKa = 4.24PTVPIPDD763 pKa = 4.38LVNTACVAGEE773 pKa = 4.31IPPQEE778 pKa = 4.43PLDD781 pKa = 3.81VPLRR785 pKa = 11.84ALAIAFDD792 pKa = 4.39EE793 pKa = 4.94EE794 pKa = 4.7PPADD798 pKa = 4.96DD799 pKa = 4.98PDD801 pKa = 4.22IDD803 pKa = 5.58GPLADD808 pKa = 5.21CDD810 pKa = 3.83EE811 pKa = 4.85ASTPVKK817 pKa = 10.52SVALDD822 pKa = 3.8GAAQCVNDD830 pKa = 4.64TPWFTYY836 pKa = 10.49AVTPYY841 pKa = 11.08NMEE844 pKa = 4.23APLPAVALIWWTLDD858 pKa = 2.86AYY860 pKa = 9.8EE861 pKa = 4.8NRR863 pKa = 11.84DD864 pKa = 3.6PSIDD868 pKa = 3.75AADD871 pKa = 3.91EE872 pKa = 4.2AALLADD878 pKa = 4.46GASQVDD884 pKa = 4.3YY885 pKa = 11.47VVIPPDD891 pKa = 3.53WAPGDD896 pKa = 3.85TLAGQQLWPGAEE908 pKa = 3.78VDD910 pKa = 4.63AEE912 pKa = 4.42GNPIDD917 pKa = 3.81WPGWTLLPDD926 pKa = 4.12GTWVLDD932 pKa = 3.74PDD934 pKa = 3.79APFYY938 pKa = 10.77DD939 pKa = 3.87LRR941 pKa = 11.84DD942 pKa = 3.54EE943 pKa = 4.35AVVEE947 pKa = 4.08IRR949 pKa = 11.84VNPSTDD955 pKa = 3.02VITAYY960 pKa = 9.85PPPTPNCNAAPPEE973 pKa = 4.22NPGPGPGPNPAGGSGSRR990 pKa = 11.84IAGTGFAGGWLVPFAVILLMGGALVAGRR1018 pKa = 11.84SWVRR1022 pKa = 11.84RR1023 pKa = 11.84RR1024 pKa = 11.84GLEE1027 pKa = 3.58QDD1029 pKa = 3.07
MM1 pKa = 7.78ANRR4 pKa = 11.84SAEE7 pKa = 3.95ASIAPPQQPLSCEE20 pKa = 3.99SLKK23 pKa = 10.85PSSEE27 pKa = 4.21LEE29 pKa = 3.98DD30 pKa = 3.06RR31 pKa = 11.84TAYY34 pKa = 10.63LDD36 pKa = 4.0CLASQNAGLRR46 pKa = 11.84SFAAAAAVDD55 pKa = 4.19EE56 pKa = 4.96NGNPDD61 pKa = 4.18LPDD64 pKa = 3.43SCGIDD69 pKa = 2.84IALILDD75 pKa = 3.52KK76 pKa = 11.11SGSIGGAGIANLKK89 pKa = 9.57SAANAFTGALVDD101 pKa = 4.53LGSQVSVVSFDD112 pKa = 3.06QDD114 pKa = 3.21AYY116 pKa = 11.58LLLPATDD123 pKa = 3.79LTSGNLGTIQGSYY136 pKa = 10.59AGLVSDD142 pKa = 5.52GWTNWKK148 pKa = 10.23RR149 pKa = 11.84GLEE152 pKa = 4.1VGLGTFGGFDD162 pKa = 3.56DD163 pKa = 4.37TVEE166 pKa = 4.1LTVVITDD173 pKa = 3.97GNPNTVNPANGGQFPDD189 pKa = 4.56GSPGALDD196 pKa = 3.64PAVLEE201 pKa = 4.34ANAIKK206 pKa = 9.11GTPSHH211 pKa = 6.03VFVVGVGPSVSTGPIVAISGAEE233 pKa = 3.82VLAADD238 pKa = 4.81GSNINTADD246 pKa = 3.48YY247 pKa = 7.46MTTTSYY253 pKa = 10.17ATLAEE258 pKa = 4.41DD259 pKa = 3.79LRR261 pKa = 11.84AVATALCGGTVVVHH275 pKa = 6.35KK276 pKa = 10.81SVDD279 pKa = 3.57GVATGGWGFEE289 pKa = 4.35TPTTDD294 pKa = 3.09VTPAALTTLADD305 pKa = 3.4GTGIFEE311 pKa = 4.36VEE313 pKa = 4.62GYY315 pKa = 9.88DD316 pKa = 3.25AQTDD320 pKa = 3.7VRR322 pKa = 11.84TVTITEE328 pKa = 4.03EE329 pKa = 3.83TRR331 pKa = 11.84PNHH334 pKa = 5.61GLEE337 pKa = 4.06NLYY340 pKa = 10.96CEE342 pKa = 4.97LNGEE346 pKa = 4.35QVPITHH352 pKa = 7.64DD353 pKa = 3.45EE354 pKa = 4.42GSPTWSVDD362 pKa = 3.14VGKK365 pKa = 10.76LDD367 pKa = 4.9IVHH370 pKa = 6.47CWVEE374 pKa = 3.9NEE376 pKa = 4.01TAKK379 pKa = 10.63WEE381 pKa = 4.22VTKK384 pKa = 10.73TSDD387 pKa = 3.86PPSGTAVEE395 pKa = 4.51AGDD398 pKa = 4.2FIDD401 pKa = 3.77YY402 pKa = 8.59TLHH405 pKa = 6.09IRR407 pKa = 11.84HH408 pKa = 6.4LSGPNVTDD416 pKa = 4.33FDD418 pKa = 4.37IDD420 pKa = 4.29DD421 pKa = 6.01DD422 pKa = 4.23ISQLAPYY429 pKa = 10.17VSFVGLLTPAPVVSSDD445 pKa = 2.76WDD447 pKa = 3.76GTPGKK452 pKa = 10.3YY453 pKa = 9.94LLKK456 pKa = 10.48LAQLTKK462 pKa = 10.91AEE464 pKa = 4.32TLNIRR469 pKa = 11.84YY470 pKa = 8.81RR471 pKa = 11.84VQVDD475 pKa = 3.02ADD477 pKa = 3.91VPPGSVLRR485 pKa = 11.84NVVLTNCPAPAEE497 pKa = 4.46GEE499 pKa = 3.83PDD501 pKa = 3.3LCVTEE506 pKa = 4.62NPTPGWEE513 pKa = 3.83LRR515 pKa = 11.84KK516 pKa = 10.17YY517 pKa = 10.52SNPPSGSVVYY527 pKa = 9.78PGYY530 pKa = 10.59EE531 pKa = 3.67VTYY534 pKa = 10.51EE535 pKa = 3.58LDD537 pKa = 3.31AYY539 pKa = 10.3NFSEE543 pKa = 4.48APVIGGIATDD553 pKa = 4.37DD554 pKa = 4.11LSDD557 pKa = 3.7VLDD560 pKa = 3.88NATIDD565 pKa = 3.63EE566 pKa = 5.07LGPGLTLEE574 pKa = 4.54GTTLTWAVPDD584 pKa = 4.67LEE586 pKa = 6.13PGDD589 pKa = 4.43SVSTSYY595 pKa = 10.46TVIVNDD601 pKa = 4.49DD602 pKa = 2.94AWGQTLTNVVTPEE615 pKa = 4.19YY616 pKa = 9.83PGSCPPPPQDD626 pKa = 3.34GEE628 pKa = 4.7VVVLAEE634 pKa = 4.59GEE636 pKa = 4.48CTTDD640 pKa = 5.03HH641 pKa = 6.61EE642 pKa = 4.81VADD645 pKa = 4.27PNLRR649 pKa = 11.84IQKK652 pKa = 9.61LWEE655 pKa = 4.38HH656 pKa = 7.23DD657 pKa = 3.76GGDD660 pKa = 3.73NPVDD664 pKa = 4.18SGDD667 pKa = 4.53DD668 pKa = 3.62PPDD671 pKa = 3.86VISYY675 pKa = 10.72AIGVWNSGNAPVEE688 pKa = 4.31NPVVTDD694 pKa = 3.65TLPEE698 pKa = 4.2GLTFVPGSEE707 pKa = 4.19VVPAGWVLDD716 pKa = 3.8ASVPGQVTFSQEE728 pKa = 3.85TPGPFDD734 pKa = 5.11PIAYY738 pKa = 8.49PGVIFEE744 pKa = 4.72FDD746 pKa = 3.73VEE748 pKa = 4.45VGEE751 pKa = 5.0LAQPDD756 pKa = 4.24PTVPIPDD763 pKa = 4.38LVNTACVAGEE773 pKa = 4.31IPPQEE778 pKa = 4.43PLDD781 pKa = 3.81VPLRR785 pKa = 11.84ALAIAFDD792 pKa = 4.39EE793 pKa = 4.94EE794 pKa = 4.7PPADD798 pKa = 4.96DD799 pKa = 4.98PDD801 pKa = 4.22IDD803 pKa = 5.58GPLADD808 pKa = 5.21CDD810 pKa = 3.83EE811 pKa = 4.85ASTPVKK817 pKa = 10.52SVALDD822 pKa = 3.8GAAQCVNDD830 pKa = 4.64TPWFTYY836 pKa = 10.49AVTPYY841 pKa = 11.08NMEE844 pKa = 4.23APLPAVALIWWTLDD858 pKa = 2.86AYY860 pKa = 9.8EE861 pKa = 4.8NRR863 pKa = 11.84DD864 pKa = 3.6PSIDD868 pKa = 3.75AADD871 pKa = 3.91EE872 pKa = 4.2AALLADD878 pKa = 4.46GASQVDD884 pKa = 4.3YY885 pKa = 11.47VVIPPDD891 pKa = 3.53WAPGDD896 pKa = 3.85TLAGQQLWPGAEE908 pKa = 3.78VDD910 pKa = 4.63AEE912 pKa = 4.42GNPIDD917 pKa = 3.81WPGWTLLPDD926 pKa = 4.12GTWVLDD932 pKa = 3.74PDD934 pKa = 3.79APFYY938 pKa = 10.77DD939 pKa = 3.87LRR941 pKa = 11.84DD942 pKa = 3.54EE943 pKa = 4.35AVVEE947 pKa = 4.08IRR949 pKa = 11.84VNPSTDD955 pKa = 3.02VITAYY960 pKa = 9.85PPPTPNCNAAPPEE973 pKa = 4.22NPGPGPGPNPAGGSGSRR990 pKa = 11.84IAGTGFAGGWLVPFAVILLMGGALVAGRR1018 pKa = 11.84SWVRR1022 pKa = 11.84RR1023 pKa = 11.84RR1024 pKa = 11.84GLEE1027 pKa = 3.58QDD1029 pKa = 3.07
Molecular weight: 107.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C1YDK2|A0A5C1YDK2_9MICO Protein translocase subunit SecA OS=Agromyces sp. KACC 19306 OX=2592652 GN=secA PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1141035 |
32 |
3214 |
340.7 |
36.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.274 ± 0.068 | 0.466 ± 0.01 |
6.422 ± 0.035 | 5.719 ± 0.037 |
3.125 ± 0.024 | 9.182 ± 0.036 |
1.94 ± 0.017 | 4.295 ± 0.03 |
1.628 ± 0.029 | 10.006 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.566 ± 0.015 | 1.785 ± 0.024 |
5.635 ± 0.031 | 2.57 ± 0.022 |
7.654 ± 0.051 | 5.276 ± 0.025 |
5.774 ± 0.038 | 9.223 ± 0.046 |
1.556 ± 0.02 | 1.903 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
