
Wuhan Insect virus 7
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
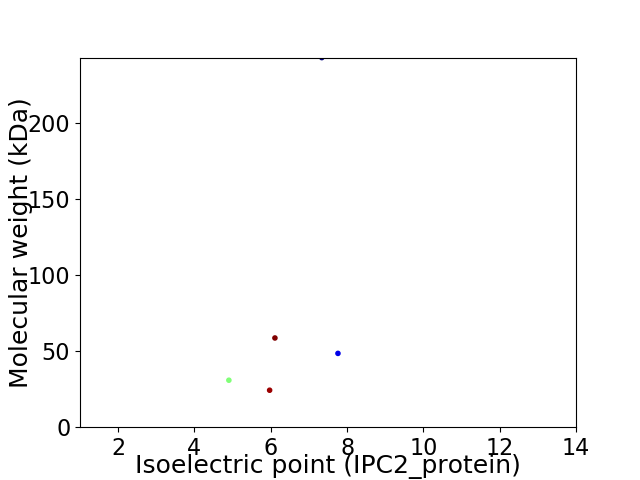
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0B5KXP4|A0A0B5KXP4_9RHAB Putative phosphoprotein OS=Wuhan Insect virus 7 OX=1608112 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.26KK2 pKa = 9.48QQNYY6 pKa = 9.31IYY8 pKa = 10.33PSMEE12 pKa = 3.83LRR14 pKa = 11.84NLTGPKK20 pKa = 10.05KK21 pKa = 10.3FFASGKK27 pKa = 9.84GYY29 pKa = 8.49LTKK32 pKa = 10.24EE33 pKa = 3.9YY34 pKa = 10.09RR35 pKa = 11.84NSPFAEE41 pKa = 3.97QGQLAVATNPRR52 pKa = 11.84RR53 pKa = 11.84TSWAMSDD60 pKa = 3.43EE61 pKa = 4.24EE62 pKa = 5.54GNIKK66 pKa = 10.61DD67 pKa = 5.2LGDD70 pKa = 3.83AFQKK74 pKa = 10.55EE75 pKa = 4.7LEE77 pKa = 4.37QKK79 pKa = 10.43HH80 pKa = 6.43SSDD83 pKa = 3.76SDD85 pKa = 3.73SQTSVSSNDD94 pKa = 3.59TVKK97 pKa = 9.97MDD99 pKa = 4.99PPPPPPPEE107 pKa = 3.66EE108 pKa = 4.48DD109 pKa = 3.3MIEE112 pKa = 4.02LVLPYY117 pKa = 9.65MEE119 pKa = 5.6IDD121 pKa = 3.79LARR124 pKa = 11.84KK125 pKa = 6.68VTEE128 pKa = 5.29AIDD131 pKa = 3.9LALKK135 pKa = 10.53DD136 pKa = 4.61LIIDD140 pKa = 3.99RR141 pKa = 11.84SLPRR145 pKa = 11.84IKK147 pKa = 11.02YY148 pKa = 9.36MMVPKK153 pKa = 9.88PKK155 pKa = 9.87EE156 pKa = 3.98ATPPPPPLQPPQEE169 pKa = 4.27TSTPPMQAQPSTSMPKK185 pKa = 10.1RR186 pKa = 11.84SGPLPPTEE194 pKa = 4.01STSRR198 pKa = 11.84EE199 pKa = 3.94RR200 pKa = 11.84EE201 pKa = 3.49RR202 pKa = 11.84SALVVEE208 pKa = 5.06FMKK211 pKa = 10.78GVKK214 pKa = 9.74LVGIYY219 pKa = 10.13EE220 pKa = 4.15PEE222 pKa = 3.49QTYY225 pKa = 7.7TLKK228 pKa = 10.59IGVGGVTEE236 pKa = 4.46DD237 pKa = 5.16LINEE241 pKa = 4.07VNEE244 pKa = 4.15QGLDD248 pKa = 3.32QPTQEE253 pKa = 4.62FIEE256 pKa = 4.26NLLCRR261 pKa = 11.84LEE263 pKa = 4.18YY264 pKa = 10.83GQTLHH269 pKa = 6.94SLYY272 pKa = 10.81EE273 pKa = 3.98LAA275 pKa = 5.75
MM1 pKa = 7.26KK2 pKa = 9.48QQNYY6 pKa = 9.31IYY8 pKa = 10.33PSMEE12 pKa = 3.83LRR14 pKa = 11.84NLTGPKK20 pKa = 10.05KK21 pKa = 10.3FFASGKK27 pKa = 9.84GYY29 pKa = 8.49LTKK32 pKa = 10.24EE33 pKa = 3.9YY34 pKa = 10.09RR35 pKa = 11.84NSPFAEE41 pKa = 3.97QGQLAVATNPRR52 pKa = 11.84RR53 pKa = 11.84TSWAMSDD60 pKa = 3.43EE61 pKa = 4.24EE62 pKa = 5.54GNIKK66 pKa = 10.61DD67 pKa = 5.2LGDD70 pKa = 3.83AFQKK74 pKa = 10.55EE75 pKa = 4.7LEE77 pKa = 4.37QKK79 pKa = 10.43HH80 pKa = 6.43SSDD83 pKa = 3.76SDD85 pKa = 3.73SQTSVSSNDD94 pKa = 3.59TVKK97 pKa = 9.97MDD99 pKa = 4.99PPPPPPPEE107 pKa = 3.66EE108 pKa = 4.48DD109 pKa = 3.3MIEE112 pKa = 4.02LVLPYY117 pKa = 9.65MEE119 pKa = 5.6IDD121 pKa = 3.79LARR124 pKa = 11.84KK125 pKa = 6.68VTEE128 pKa = 5.29AIDD131 pKa = 3.9LALKK135 pKa = 10.53DD136 pKa = 4.61LIIDD140 pKa = 3.99RR141 pKa = 11.84SLPRR145 pKa = 11.84IKK147 pKa = 11.02YY148 pKa = 9.36MMVPKK153 pKa = 9.88PKK155 pKa = 9.87EE156 pKa = 3.98ATPPPPPLQPPQEE169 pKa = 4.27TSTPPMQAQPSTSMPKK185 pKa = 10.1RR186 pKa = 11.84SGPLPPTEE194 pKa = 4.01STSRR198 pKa = 11.84EE199 pKa = 3.94RR200 pKa = 11.84EE201 pKa = 3.49RR202 pKa = 11.84SALVVEE208 pKa = 5.06FMKK211 pKa = 10.78GVKK214 pKa = 9.74LVGIYY219 pKa = 10.13EE220 pKa = 4.15PEE222 pKa = 3.49QTYY225 pKa = 7.7TLKK228 pKa = 10.59IGVGGVTEE236 pKa = 4.46DD237 pKa = 5.16LINEE241 pKa = 4.07VNEE244 pKa = 4.15QGLDD248 pKa = 3.32QPTQEE253 pKa = 4.62FIEE256 pKa = 4.26NLLCRR261 pKa = 11.84LEE263 pKa = 4.18YY264 pKa = 10.83GQTLHH269 pKa = 6.94SLYY272 pKa = 10.81EE273 pKa = 3.98LAA275 pKa = 5.75
Molecular weight: 30.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KXP4|A0A0B5KXP4_9RHAB Putative phosphoprotein OS=Wuhan Insect virus 7 OX=1608112 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.55SKK3 pKa = 10.3KK4 pKa = 10.33QFFKK8 pKa = 11.21VGDD11 pKa = 3.62LTRR14 pKa = 11.84TQVTLLAPHH23 pKa = 6.44PQVPAQYY30 pKa = 8.94PSEE33 pKa = 4.11YY34 pKa = 10.36FVNNKK39 pKa = 9.28NKK41 pKa = 9.65PKK43 pKa = 10.56VVLSVKK49 pKa = 10.1GLNVEE54 pKa = 4.46NIRR57 pKa = 11.84AVINSGLSNNNLRR70 pKa = 11.84LAPSLYY76 pKa = 10.46FIYY79 pKa = 10.18LYY81 pKa = 10.37FLNVKK86 pKa = 8.79TVLTKK91 pKa = 9.77KK92 pKa = 9.62WEE94 pKa = 4.11SFGIKK99 pKa = 10.12FGDD102 pKa = 3.75PNEE105 pKa = 4.42EE106 pKa = 3.83VTPWSLIDD114 pKa = 3.59VQLDD118 pKa = 3.33SDD120 pKa = 4.33SGVDD124 pKa = 3.53HH125 pKa = 7.48PSLEE129 pKa = 4.89GIAKK133 pKa = 10.19SDD135 pKa = 4.19DD136 pKa = 2.97NWIICLLLSMHH147 pKa = 6.57RR148 pKa = 11.84LKK150 pKa = 11.0SGMLDD155 pKa = 3.37SYY157 pKa = 11.54RR158 pKa = 11.84EE159 pKa = 4.09IIQKK163 pKa = 10.29RR164 pKa = 11.84IYY166 pKa = 10.66DD167 pKa = 3.37QMHH170 pKa = 5.34QYY172 pKa = 10.76GYY174 pKa = 10.96KK175 pKa = 9.99KK176 pKa = 10.42GGYY179 pKa = 8.01PSSFTSEE186 pKa = 3.82DD187 pKa = 3.07WAGSLDD193 pKa = 3.84FTRR196 pKa = 11.84MVAAIDD202 pKa = 3.57MFLSKK207 pKa = 10.62FPTLVQSQLRR217 pKa = 11.84ICTLTSRR224 pKa = 11.84FKK226 pKa = 11.16DD227 pKa = 3.5CTALMSLGFIKK238 pKa = 10.42EE239 pKa = 4.23VLCLPDD245 pKa = 3.4EE246 pKa = 4.67SEE248 pKa = 4.02FLDD251 pKa = 4.25WIFTAPIAAEE261 pKa = 4.01AEE263 pKa = 3.83AMARR267 pKa = 11.84HH268 pKa = 5.25GQEE271 pKa = 3.71TLEE274 pKa = 4.37PDD276 pKa = 3.49SYY278 pKa = 11.48FPYY281 pKa = 10.57QSDD284 pKa = 3.27LRR286 pKa = 11.84LVNRR290 pKa = 11.84THH292 pKa = 6.66YY293 pKa = 10.46SASANEE299 pKa = 4.38NIFFLIHH306 pKa = 7.01AIGSLLHH313 pKa = 5.67VQRR316 pKa = 11.84SMNAKK321 pKa = 8.5KK322 pKa = 7.45TTDD325 pKa = 3.07KK326 pKa = 11.44NIINNSCNAKK336 pKa = 9.71IIAYY340 pKa = 8.85VFSNTFKK347 pKa = 10.37MAKK350 pKa = 8.99QYY352 pKa = 10.77QAIGGPAVQKK362 pKa = 10.87RR363 pKa = 11.84NLNINKK369 pKa = 9.56PEE371 pKa = 4.22DD372 pKa = 4.2LEE374 pKa = 4.84PKK376 pKa = 8.02TQNAALWYY384 pKa = 10.08AYY386 pKa = 7.51MQSYY390 pKa = 10.48DD391 pKa = 4.19FKK393 pKa = 10.34LTPNMRR399 pKa = 11.84AFVEE403 pKa = 4.29RR404 pKa = 11.84SKK406 pKa = 11.8AEE408 pKa = 3.69MGEE411 pKa = 4.24TRR413 pKa = 11.84EE414 pKa = 4.28GTIGHH419 pKa = 5.76YY420 pKa = 8.88VQHH423 pKa = 6.7HH424 pKa = 5.56FF425 pKa = 4.63
MM1 pKa = 7.55SKK3 pKa = 10.3KK4 pKa = 10.33QFFKK8 pKa = 11.21VGDD11 pKa = 3.62LTRR14 pKa = 11.84TQVTLLAPHH23 pKa = 6.44PQVPAQYY30 pKa = 8.94PSEE33 pKa = 4.11YY34 pKa = 10.36FVNNKK39 pKa = 9.28NKK41 pKa = 9.65PKK43 pKa = 10.56VVLSVKK49 pKa = 10.1GLNVEE54 pKa = 4.46NIRR57 pKa = 11.84AVINSGLSNNNLRR70 pKa = 11.84LAPSLYY76 pKa = 10.46FIYY79 pKa = 10.18LYY81 pKa = 10.37FLNVKK86 pKa = 8.79TVLTKK91 pKa = 9.77KK92 pKa = 9.62WEE94 pKa = 4.11SFGIKK99 pKa = 10.12FGDD102 pKa = 3.75PNEE105 pKa = 4.42EE106 pKa = 3.83VTPWSLIDD114 pKa = 3.59VQLDD118 pKa = 3.33SDD120 pKa = 4.33SGVDD124 pKa = 3.53HH125 pKa = 7.48PSLEE129 pKa = 4.89GIAKK133 pKa = 10.19SDD135 pKa = 4.19DD136 pKa = 2.97NWIICLLLSMHH147 pKa = 6.57RR148 pKa = 11.84LKK150 pKa = 11.0SGMLDD155 pKa = 3.37SYY157 pKa = 11.54RR158 pKa = 11.84EE159 pKa = 4.09IIQKK163 pKa = 10.29RR164 pKa = 11.84IYY166 pKa = 10.66DD167 pKa = 3.37QMHH170 pKa = 5.34QYY172 pKa = 10.76GYY174 pKa = 10.96KK175 pKa = 9.99KK176 pKa = 10.42GGYY179 pKa = 8.01PSSFTSEE186 pKa = 3.82DD187 pKa = 3.07WAGSLDD193 pKa = 3.84FTRR196 pKa = 11.84MVAAIDD202 pKa = 3.57MFLSKK207 pKa = 10.62FPTLVQSQLRR217 pKa = 11.84ICTLTSRR224 pKa = 11.84FKK226 pKa = 11.16DD227 pKa = 3.5CTALMSLGFIKK238 pKa = 10.42EE239 pKa = 4.23VLCLPDD245 pKa = 3.4EE246 pKa = 4.67SEE248 pKa = 4.02FLDD251 pKa = 4.25WIFTAPIAAEE261 pKa = 4.01AEE263 pKa = 3.83AMARR267 pKa = 11.84HH268 pKa = 5.25GQEE271 pKa = 3.71TLEE274 pKa = 4.37PDD276 pKa = 3.49SYY278 pKa = 11.48FPYY281 pKa = 10.57QSDD284 pKa = 3.27LRR286 pKa = 11.84LVNRR290 pKa = 11.84THH292 pKa = 6.66YY293 pKa = 10.46SASANEE299 pKa = 4.38NIFFLIHH306 pKa = 7.01AIGSLLHH313 pKa = 5.67VQRR316 pKa = 11.84SMNAKK321 pKa = 8.5KK322 pKa = 7.45TTDD325 pKa = 3.07KK326 pKa = 11.44NIINNSCNAKK336 pKa = 9.71IIAYY340 pKa = 8.85VFSNTFKK347 pKa = 10.37MAKK350 pKa = 8.99QYY352 pKa = 10.77QAIGGPAVQKK362 pKa = 10.87RR363 pKa = 11.84NLNINKK369 pKa = 9.56PEE371 pKa = 4.22DD372 pKa = 4.2LEE374 pKa = 4.84PKK376 pKa = 8.02TQNAALWYY384 pKa = 10.08AYY386 pKa = 7.51MQSYY390 pKa = 10.48DD391 pKa = 4.19FKK393 pKa = 10.34LTPNMRR399 pKa = 11.84AFVEE403 pKa = 4.29RR404 pKa = 11.84SKK406 pKa = 11.8AEE408 pKa = 3.69MGEE411 pKa = 4.24TRR413 pKa = 11.84EE414 pKa = 4.28GTIGHH419 pKa = 5.76YY420 pKa = 8.88VQHH423 pKa = 6.7HH424 pKa = 5.56FF425 pKa = 4.63
Molecular weight: 48.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3554 |
212 |
2120 |
710.8 |
81.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.361 ± 0.562 | 1.716 ± 0.48 |
5.29 ± 0.099 | 5.881 ± 0.647 |
4.221 ± 0.35 | 5.346 ± 0.142 |
2.673 ± 0.461 | 6.669 ± 0.425 |
6.021 ± 0.269 | 10.298 ± 0.686 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.011 ± 0.221 | 4.699 ± 0.336 |
5.627 ± 0.965 | 3.658 ± 0.416 |
5.093 ± 0.473 | 8.666 ± 0.245 |
5.515 ± 0.159 | 5.824 ± 0.76 |
1.66 ± 0.225 | 3.77 ± 0.216 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
