
Chenuda virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 7.5
Get precalculated fractions of proteins
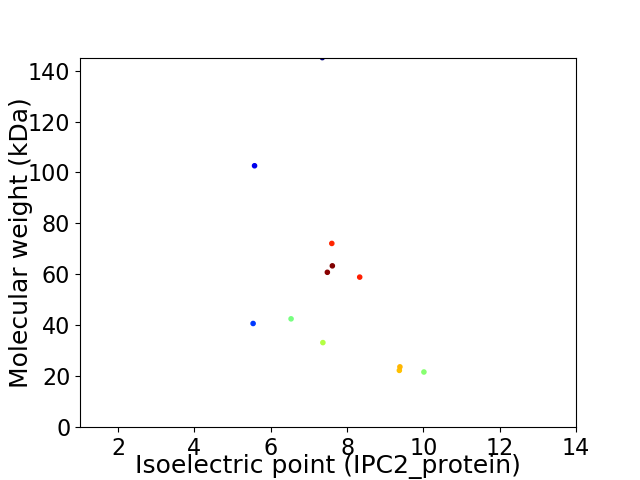
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
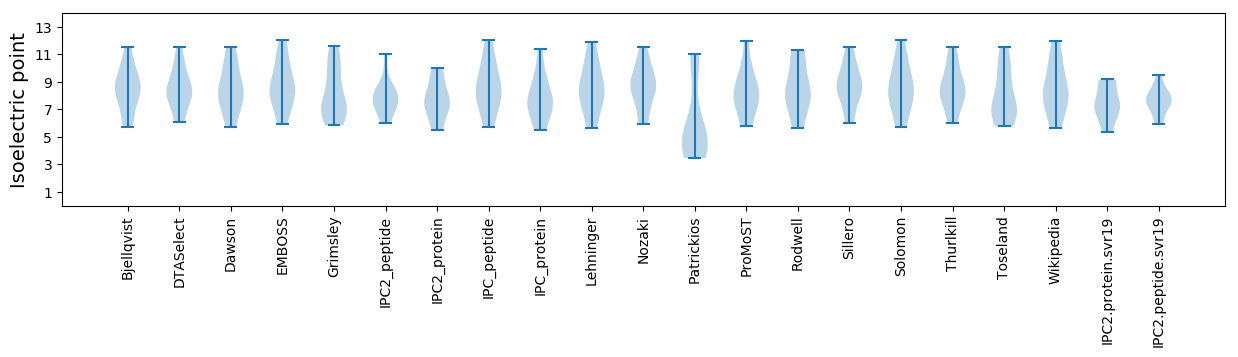
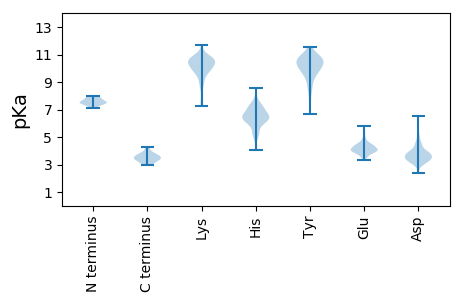
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4MKD1|A0A0H4MKD1_9REOV Core protein VP7 OS=Chenuda virus OX=40065 GN=VP7 PE=3 SV=1
MM1 pKa = 7.89DD2 pKa = 5.84AYY4 pKa = 10.62NARR7 pKa = 11.84ALSVLEE13 pKa = 4.05GLALAADD20 pKa = 4.33PRR22 pKa = 11.84AHH24 pKa = 7.34RR25 pKa = 11.84DD26 pKa = 3.47PVSEE30 pKa = 4.18TTLSIFMMRR39 pKa = 11.84FNSTTTRR46 pKa = 11.84PIVGAPTTRR55 pKa = 11.84EE56 pKa = 3.42ARR58 pKa = 11.84RR59 pKa = 11.84NNFYY63 pKa = 10.95AALDD67 pKa = 3.52VAYY70 pKa = 9.41AALGITSQFLMPGYY84 pKa = 9.49VQNPQTLAILARR96 pKa = 11.84DD97 pKa = 4.53EE98 pKa = 4.44IPYY101 pKa = 9.29TPSTFRR107 pKa = 11.84RR108 pKa = 11.84VQRR111 pKa = 11.84IRR113 pKa = 11.84ICSEE117 pKa = 3.82GACTFRR123 pKa = 11.84EE124 pKa = 4.54EE125 pKa = 4.06YY126 pKa = 10.43FPYY129 pKa = 10.43QNYY132 pKa = 8.5TMLTGRR138 pKa = 11.84AMPINPPADD147 pKa = 3.9GGPRR151 pKa = 11.84CYY153 pKa = 10.45LVDD156 pKa = 4.16PNTVQVHH163 pKa = 5.21VEE165 pKa = 4.04PEE167 pKa = 3.59QTIAVTDD174 pKa = 3.86VVVPTAPNWVAAEE187 pKa = 4.07LAWYY191 pKa = 8.89IQTNGPGANAAEE203 pKa = 5.69GYY205 pKa = 10.1AQDD208 pKa = 2.92VDD210 pKa = 4.72VYY212 pKa = 11.37VNDD215 pKa = 3.47RR216 pKa = 11.84RR217 pKa = 11.84LPAGLPYY224 pKa = 10.58RR225 pKa = 11.84LNPGDD230 pKa = 4.0RR231 pKa = 11.84VEE233 pKa = 4.62LTNHH237 pKa = 7.21DD238 pKa = 4.35DD239 pKa = 4.1FNLGTAMFFIRR250 pKa = 11.84RR251 pKa = 11.84YY252 pKa = 8.65WSAAPPQVIYY262 pKa = 11.04DD263 pKa = 3.72SMEE266 pKa = 4.65ADD268 pKa = 2.79ICAVYY273 pKa = 9.93IYY275 pKa = 10.46HH276 pKa = 7.49DD277 pKa = 4.18RR278 pKa = 11.84TWHH281 pKa = 6.4RR282 pKa = 11.84LRR284 pKa = 11.84SYY286 pKa = 10.44ICTQVGLPPTYY297 pKa = 10.08YY298 pKa = 9.9PSEE301 pKa = 4.07ATQEE305 pKa = 3.94PRR307 pKa = 11.84RR308 pKa = 11.84VLTIAILSRR317 pKa = 11.84LFDD320 pKa = 3.73VYY322 pKa = 10.84CALSPEE328 pKa = 3.97IHH330 pKa = 6.56LPAPEE335 pKa = 4.14VAPGDD340 pKa = 3.39LAQRR344 pKa = 11.84LRR346 pKa = 11.84EE347 pKa = 4.04ALQTLRR353 pKa = 11.84GAPPPAAPRR362 pKa = 11.84GGQQ365 pKa = 2.99
MM1 pKa = 7.89DD2 pKa = 5.84AYY4 pKa = 10.62NARR7 pKa = 11.84ALSVLEE13 pKa = 4.05GLALAADD20 pKa = 4.33PRR22 pKa = 11.84AHH24 pKa = 7.34RR25 pKa = 11.84DD26 pKa = 3.47PVSEE30 pKa = 4.18TTLSIFMMRR39 pKa = 11.84FNSTTTRR46 pKa = 11.84PIVGAPTTRR55 pKa = 11.84EE56 pKa = 3.42ARR58 pKa = 11.84RR59 pKa = 11.84NNFYY63 pKa = 10.95AALDD67 pKa = 3.52VAYY70 pKa = 9.41AALGITSQFLMPGYY84 pKa = 9.49VQNPQTLAILARR96 pKa = 11.84DD97 pKa = 4.53EE98 pKa = 4.44IPYY101 pKa = 9.29TPSTFRR107 pKa = 11.84RR108 pKa = 11.84VQRR111 pKa = 11.84IRR113 pKa = 11.84ICSEE117 pKa = 3.82GACTFRR123 pKa = 11.84EE124 pKa = 4.54EE125 pKa = 4.06YY126 pKa = 10.43FPYY129 pKa = 10.43QNYY132 pKa = 8.5TMLTGRR138 pKa = 11.84AMPINPPADD147 pKa = 3.9GGPRR151 pKa = 11.84CYY153 pKa = 10.45LVDD156 pKa = 4.16PNTVQVHH163 pKa = 5.21VEE165 pKa = 4.04PEE167 pKa = 3.59QTIAVTDD174 pKa = 3.86VVVPTAPNWVAAEE187 pKa = 4.07LAWYY191 pKa = 8.89IQTNGPGANAAEE203 pKa = 5.69GYY205 pKa = 10.1AQDD208 pKa = 2.92VDD210 pKa = 4.72VYY212 pKa = 11.37VNDD215 pKa = 3.47RR216 pKa = 11.84RR217 pKa = 11.84LPAGLPYY224 pKa = 10.58RR225 pKa = 11.84LNPGDD230 pKa = 4.0RR231 pKa = 11.84VEE233 pKa = 4.62LTNHH237 pKa = 7.21DD238 pKa = 4.35DD239 pKa = 4.1FNLGTAMFFIRR250 pKa = 11.84RR251 pKa = 11.84YY252 pKa = 8.65WSAAPPQVIYY262 pKa = 11.04DD263 pKa = 3.72SMEE266 pKa = 4.65ADD268 pKa = 2.79ICAVYY273 pKa = 9.93IYY275 pKa = 10.46HH276 pKa = 7.49DD277 pKa = 4.18RR278 pKa = 11.84TWHH281 pKa = 6.4RR282 pKa = 11.84LRR284 pKa = 11.84SYY286 pKa = 10.44ICTQVGLPPTYY297 pKa = 10.08YY298 pKa = 9.9PSEE301 pKa = 4.07ATQEE305 pKa = 3.94PRR307 pKa = 11.84RR308 pKa = 11.84VLTIAILSRR317 pKa = 11.84LFDD320 pKa = 3.73VYY322 pKa = 10.84CALSPEE328 pKa = 3.97IHH330 pKa = 6.56LPAPEE335 pKa = 4.14VAPGDD340 pKa = 3.39LAQRR344 pKa = 11.84LRR346 pKa = 11.84EE347 pKa = 4.04ALQTLRR353 pKa = 11.84GAPPPAAPRR362 pKa = 11.84GGQQ365 pKa = 2.99
Molecular weight: 40.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4M927|A0A0H4M927_9REOV Outer capsid protein OS=Chenuda virus OX=40065 GN=VP4 PE=4 SV=1
MM1 pKa = 7.53LSVADD6 pKa = 3.96AKK8 pKa = 10.7RR9 pKa = 11.84RR10 pKa = 11.84EE11 pKa = 4.16QMAGTEE17 pKa = 4.07MRR19 pKa = 11.84PLVPSAPRR27 pKa = 11.84PLPGAMPEE35 pKa = 3.94MALGVLQNALTSSTGANEE53 pKa = 3.75TAKK56 pKa = 10.74NEE58 pKa = 3.88KK59 pKa = 9.6AAYY62 pKa = 8.52GAASEE67 pKa = 4.67VMRR70 pKa = 11.84DD71 pKa = 3.74DD72 pKa = 3.95PATRR76 pKa = 11.84QLKK79 pKa = 8.69VHH81 pKa = 6.07VSVTAIAEE89 pKa = 3.95LEE91 pKa = 3.93EE92 pKa = 4.28RR93 pKa = 11.84YY94 pKa = 10.29KK95 pKa = 10.36RR96 pKa = 11.84VRR98 pKa = 11.84RR99 pKa = 11.84KK100 pKa = 9.97KK101 pKa = 10.23CACKK105 pKa = 9.17WLQAISSALVLTTSIAMSIVTASTWVEE132 pKa = 3.58NTLVKK137 pKa = 10.81NGVPVYY143 pKa = 9.47MIISGLTTVAMVAGRR158 pKa = 11.84MHH160 pKa = 6.59AHH162 pKa = 6.58ARR164 pKa = 11.84TATRR168 pKa = 11.84TIKK171 pKa = 10.37RR172 pKa = 11.84DD173 pKa = 3.41LVKK176 pKa = 10.6KK177 pKa = 10.53KK178 pKa = 10.71SYY180 pKa = 10.02VQLAVAMGTPLPGTVAAPAKK200 pKa = 10.36AGTSVGAASIVDD212 pKa = 3.75DD213 pKa = 4.33LVARR217 pKa = 11.84GWQPLPP223 pKa = 3.77
MM1 pKa = 7.53LSVADD6 pKa = 3.96AKK8 pKa = 10.7RR9 pKa = 11.84RR10 pKa = 11.84EE11 pKa = 4.16QMAGTEE17 pKa = 4.07MRR19 pKa = 11.84PLVPSAPRR27 pKa = 11.84PLPGAMPEE35 pKa = 3.94MALGVLQNALTSSTGANEE53 pKa = 3.75TAKK56 pKa = 10.74NEE58 pKa = 3.88KK59 pKa = 9.6AAYY62 pKa = 8.52GAASEE67 pKa = 4.67VMRR70 pKa = 11.84DD71 pKa = 3.74DD72 pKa = 3.95PATRR76 pKa = 11.84QLKK79 pKa = 8.69VHH81 pKa = 6.07VSVTAIAEE89 pKa = 3.95LEE91 pKa = 3.93EE92 pKa = 4.28RR93 pKa = 11.84YY94 pKa = 10.29KK95 pKa = 10.36RR96 pKa = 11.84VRR98 pKa = 11.84RR99 pKa = 11.84KK100 pKa = 9.97KK101 pKa = 10.23CACKK105 pKa = 9.17WLQAISSALVLTTSIAMSIVTASTWVEE132 pKa = 3.58NTLVKK137 pKa = 10.81NGVPVYY143 pKa = 9.47MIISGLTTVAMVAGRR158 pKa = 11.84MHH160 pKa = 6.59AHH162 pKa = 6.58ARR164 pKa = 11.84TATRR168 pKa = 11.84TIKK171 pKa = 10.37RR172 pKa = 11.84DD173 pKa = 3.41LVKK176 pKa = 10.6KK177 pKa = 10.53KK178 pKa = 10.71SYY180 pKa = 10.02VQLAVAMGTPLPGTVAAPAKK200 pKa = 10.36AGTSVGAASIVDD212 pKa = 3.75DD213 pKa = 4.33LVARR217 pKa = 11.84GWQPLPP223 pKa = 3.77
Molecular weight: 23.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6145 |
183 |
1285 |
512.1 |
57.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.301 ± 0.604 | 1.383 ± 0.242 |
5.435 ± 0.341 | 5.891 ± 0.249 |
3.466 ± 0.333 | 5.696 ± 0.395 |
2.571 ± 0.226 | 5.028 ± 0.275 |
2.799 ± 0.503 | 9.552 ± 0.362 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.457 ± 0.264 | 2.587 ± 0.212 |
6.151 ± 0.427 | 3.531 ± 0.417 |
9.39 ± 0.431 | 5.663 ± 0.391 |
6.363 ± 0.37 | 7.681 ± 0.324 |
1.155 ± 0.187 | 2.897 ± 0.276 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
