
Acidisphaera rubrifaciens HS-AP3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Acidisphaera; Acidisphaera rubrifaciens
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
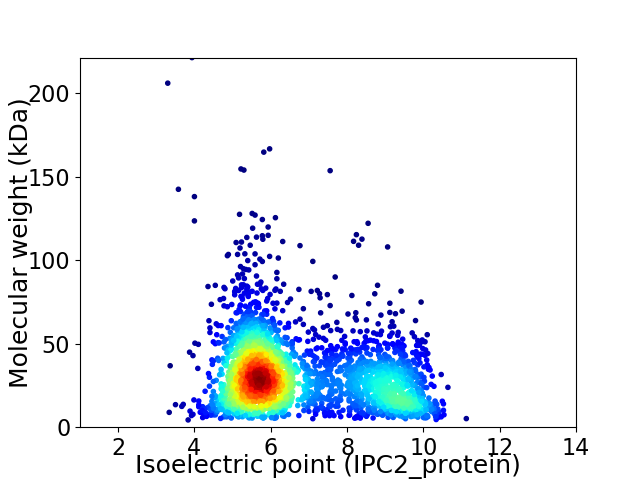
Virtual 2D-PAGE plot for 2788 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D6P945|A0A0D6P945_9PROT Transcriptional regulator polyhydroxyalkanoate biosynthesis OS=Acidisphaera rubrifaciens HS-AP3 OX=1231350 GN=Asru_0493_03 PE=4 SV=1
MM1 pKa = 7.5ANAVTYY7 pKa = 10.31LPSFPQPTSPNQIVGLILQNNSGTATPAQNLTFGEE42 pKa = 4.46VFAAGAVASGANLVAQIGGSATPVQMDD69 pKa = 3.63VKK71 pKa = 10.02STYY74 pKa = 10.96ADD76 pKa = 3.36GSVEE80 pKa = 4.27SAVLTVAAPTLAANSSTPVMLSLGGSAPAPAVNLAGALSNYY121 pKa = 10.21SLTAALMVNGTAYY134 pKa = 10.98NIDD137 pKa = 4.11LTSAMQTALANGTANYY153 pKa = 8.12WLKK156 pKa = 11.24GPDD159 pKa = 3.02ATQARR164 pKa = 11.84VSVPVAGSLRR174 pKa = 11.84LVADD178 pKa = 4.07ITANADD184 pKa = 3.36GTFSATVGFNNDD196 pKa = 2.56VAEE199 pKa = 4.22TANGGTIALNAKK211 pKa = 9.75LIQNGSTVYY220 pKa = 10.64SSGNITQYY228 pKa = 10.94QYY230 pKa = 11.38QDD232 pKa = 2.52WSATVGTAPASTINVQHH249 pKa = 7.49DD250 pKa = 2.88IGYY253 pKa = 9.83LEE255 pKa = 4.32KK256 pKa = 10.26TGAIPTYY263 pKa = 10.52DD264 pKa = 3.8LSTGVATSVLTGDD277 pKa = 4.06ASAVASPGWNAPLAPNGINQHH298 pKa = 5.58MPTTGGRR305 pKa = 11.84GDD307 pKa = 3.85IGPTTQANADD317 pKa = 3.51WLQTQDD323 pKa = 3.38QSAMQYY329 pKa = 11.81ALGQAGNGASSVPWHH344 pKa = 6.47FWNPTAGTWLNTASYY359 pKa = 8.36PQLWNDD365 pKa = 3.19PRR367 pKa = 11.84GGNGAPGGLTQQVDD381 pKa = 3.56TGNTGWTVDD390 pKa = 3.72EE391 pKa = 4.3AHH393 pKa = 6.46QPTLDD398 pKa = 3.37YY399 pKa = 11.03DD400 pKa = 4.14AYY402 pKa = 11.17LLTGNRR408 pKa = 11.84TYY410 pKa = 11.44LDD412 pKa = 3.6NLNAQASNAVMADD425 pKa = 3.13WPGVRR430 pKa = 11.84DD431 pKa = 3.52EE432 pKa = 5.16GSYY435 pKa = 10.89TDD437 pKa = 3.59IVANGGDD444 pKa = 3.83QVRR447 pKa = 11.84QQAWSLRR454 pKa = 11.84EE455 pKa = 3.7IDD457 pKa = 3.33EE458 pKa = 4.28AAYY461 pKa = 10.5ANPAGSAEE469 pKa = 4.08KK470 pKa = 10.39AYY472 pKa = 8.42FTQTMNDD479 pKa = 2.83NWHH482 pKa = 6.45WLVSQIPAWTKK493 pKa = 11.16LEE495 pKa = 4.06GQAYY499 pKa = 9.17GYY501 pKa = 11.13VPGTYY506 pKa = 10.16GSTGTTMAPWQQDD519 pKa = 3.75YY520 pKa = 10.25FVSTTVEE527 pKa = 3.82AARR530 pKa = 11.84MGNADD535 pKa = 4.32ALTFLKK541 pKa = 10.27WEE543 pKa = 4.58SNFIIGRR550 pKa = 11.84FSQSQPGWNAHH561 pKa = 6.98DD562 pKa = 3.79GAAYY566 pKa = 10.74NLDD569 pKa = 3.52IGPANSPLQTWSAIEE584 pKa = 4.17SATAAAGDD592 pKa = 4.27SNGSGWAAGDD602 pKa = 3.7YY603 pKa = 10.58AQLGLEE609 pKa = 4.2SLAGIYY615 pKa = 10.34SVTGDD620 pKa = 3.68PQALAEE626 pKa = 4.21NNWLKK631 pKa = 11.13SSGAPFLDD639 pKa = 3.56TASYY643 pKa = 10.27QGLGSSPAEE652 pKa = 4.05QFNVAPTPSTLPPSGAPAPTPTPTPTPTPTPTPTPTPAPTPTPTPTPTTNPTPVPSSGHH711 pKa = 4.53VLQVGAGQTYY721 pKa = 8.1ATISQAVAASQSGDD735 pKa = 3.71TIQVQAGTYY744 pKa = 9.87TNDD747 pKa = 3.08FPNLISHH754 pKa = 7.58DD755 pKa = 4.03LTLEE759 pKa = 4.0AVGGVVHH766 pKa = 6.19MVATEE771 pKa = 3.71PVPNGKK777 pKa = 10.21AILDD781 pKa = 3.42IGAPGVSVSVTGFDD795 pKa = 3.61FSGATVSSGNGAGIRR810 pKa = 11.84YY811 pKa = 9.18EE812 pKa = 4.37GGSLTLNNDD821 pKa = 3.11SFEE824 pKa = 4.47NNQDD828 pKa = 3.54GLLGASDD835 pKa = 3.59TSGAITITNSVFSHH849 pKa = 6.39NGTGDD854 pKa = 3.34GHH856 pKa = 4.99THH858 pKa = 5.77NLYY861 pKa = 10.34VGEE864 pKa = 4.32VGTLTVTGSTLTDD877 pKa = 2.9AVVGHH882 pKa = 6.56EE883 pKa = 4.1IKK885 pKa = 10.76SRR887 pKa = 11.84ADD889 pKa = 3.15TTIIQGNTISDD900 pKa = 4.07GPTGTGSYY908 pKa = 10.59SIDD911 pKa = 3.56LPNGGKK917 pKa = 9.93AVISGNVIEE926 pKa = 5.11KK927 pKa = 10.5GPNAGNPAIISVGEE941 pKa = 3.97EE942 pKa = 3.7GGVYY946 pKa = 10.52ANSSVTLSNNTVLNDD961 pKa = 3.45NASPSVVLVKK971 pKa = 10.82NDD973 pKa = 3.42TTGPATISGTSVWNLTASEE992 pKa = 4.18MLQGPGSVSGTTLLASEE1009 pKa = 4.66PSIAAAAAPTPTPTPTPAPTPTPTPTPTPADD1040 pKa = 3.77TPIVVAPGGTLTTAAGTTNTVTMPTGDD1067 pKa = 3.33ATVVSGGTDD1076 pKa = 3.55TIYY1079 pKa = 11.04AGAGSDD1085 pKa = 3.95KK1086 pKa = 10.56ISSGVNTVTVNQGSGPLTFIGGYY1109 pKa = 9.55GGNATLNMAAGGTGANLTLYY1129 pKa = 11.14GNTTINAAGTFTANDD1144 pKa = 3.92GGSGAGADD1152 pKa = 3.55VFNITKK1158 pKa = 10.59GDD1160 pKa = 3.58VTNITITNFGDD1171 pKa = 3.56AFHH1174 pKa = 7.36LIGFSAGEE1182 pKa = 3.93AQQAVATAQHH1192 pKa = 5.99TGAGEE1197 pKa = 4.08VMSFSDD1203 pKa = 4.63GSQVTLAGWNHH1214 pKa = 6.31AMLSWFQQ1221 pKa = 3.24
MM1 pKa = 7.5ANAVTYY7 pKa = 10.31LPSFPQPTSPNQIVGLILQNNSGTATPAQNLTFGEE42 pKa = 4.46VFAAGAVASGANLVAQIGGSATPVQMDD69 pKa = 3.63VKK71 pKa = 10.02STYY74 pKa = 10.96ADD76 pKa = 3.36GSVEE80 pKa = 4.27SAVLTVAAPTLAANSSTPVMLSLGGSAPAPAVNLAGALSNYY121 pKa = 10.21SLTAALMVNGTAYY134 pKa = 10.98NIDD137 pKa = 4.11LTSAMQTALANGTANYY153 pKa = 8.12WLKK156 pKa = 11.24GPDD159 pKa = 3.02ATQARR164 pKa = 11.84VSVPVAGSLRR174 pKa = 11.84LVADD178 pKa = 4.07ITANADD184 pKa = 3.36GTFSATVGFNNDD196 pKa = 2.56VAEE199 pKa = 4.22TANGGTIALNAKK211 pKa = 9.75LIQNGSTVYY220 pKa = 10.64SSGNITQYY228 pKa = 10.94QYY230 pKa = 11.38QDD232 pKa = 2.52WSATVGTAPASTINVQHH249 pKa = 7.49DD250 pKa = 2.88IGYY253 pKa = 9.83LEE255 pKa = 4.32KK256 pKa = 10.26TGAIPTYY263 pKa = 10.52DD264 pKa = 3.8LSTGVATSVLTGDD277 pKa = 4.06ASAVASPGWNAPLAPNGINQHH298 pKa = 5.58MPTTGGRR305 pKa = 11.84GDD307 pKa = 3.85IGPTTQANADD317 pKa = 3.51WLQTQDD323 pKa = 3.38QSAMQYY329 pKa = 11.81ALGQAGNGASSVPWHH344 pKa = 6.47FWNPTAGTWLNTASYY359 pKa = 8.36PQLWNDD365 pKa = 3.19PRR367 pKa = 11.84GGNGAPGGLTQQVDD381 pKa = 3.56TGNTGWTVDD390 pKa = 3.72EE391 pKa = 4.3AHH393 pKa = 6.46QPTLDD398 pKa = 3.37YY399 pKa = 11.03DD400 pKa = 4.14AYY402 pKa = 11.17LLTGNRR408 pKa = 11.84TYY410 pKa = 11.44LDD412 pKa = 3.6NLNAQASNAVMADD425 pKa = 3.13WPGVRR430 pKa = 11.84DD431 pKa = 3.52EE432 pKa = 5.16GSYY435 pKa = 10.89TDD437 pKa = 3.59IVANGGDD444 pKa = 3.83QVRR447 pKa = 11.84QQAWSLRR454 pKa = 11.84EE455 pKa = 3.7IDD457 pKa = 3.33EE458 pKa = 4.28AAYY461 pKa = 10.5ANPAGSAEE469 pKa = 4.08KK470 pKa = 10.39AYY472 pKa = 8.42FTQTMNDD479 pKa = 2.83NWHH482 pKa = 6.45WLVSQIPAWTKK493 pKa = 11.16LEE495 pKa = 4.06GQAYY499 pKa = 9.17GYY501 pKa = 11.13VPGTYY506 pKa = 10.16GSTGTTMAPWQQDD519 pKa = 3.75YY520 pKa = 10.25FVSTTVEE527 pKa = 3.82AARR530 pKa = 11.84MGNADD535 pKa = 4.32ALTFLKK541 pKa = 10.27WEE543 pKa = 4.58SNFIIGRR550 pKa = 11.84FSQSQPGWNAHH561 pKa = 6.98DD562 pKa = 3.79GAAYY566 pKa = 10.74NLDD569 pKa = 3.52IGPANSPLQTWSAIEE584 pKa = 4.17SATAAAGDD592 pKa = 4.27SNGSGWAAGDD602 pKa = 3.7YY603 pKa = 10.58AQLGLEE609 pKa = 4.2SLAGIYY615 pKa = 10.34SVTGDD620 pKa = 3.68PQALAEE626 pKa = 4.21NNWLKK631 pKa = 11.13SSGAPFLDD639 pKa = 3.56TASYY643 pKa = 10.27QGLGSSPAEE652 pKa = 4.05QFNVAPTPSTLPPSGAPAPTPTPTPTPTPTPTPTPTPAPTPTPTPTPTTNPTPVPSSGHH711 pKa = 4.53VLQVGAGQTYY721 pKa = 8.1ATISQAVAASQSGDD735 pKa = 3.71TIQVQAGTYY744 pKa = 9.87TNDD747 pKa = 3.08FPNLISHH754 pKa = 7.58DD755 pKa = 4.03LTLEE759 pKa = 4.0AVGGVVHH766 pKa = 6.19MVATEE771 pKa = 3.71PVPNGKK777 pKa = 10.21AILDD781 pKa = 3.42IGAPGVSVSVTGFDD795 pKa = 3.61FSGATVSSGNGAGIRR810 pKa = 11.84YY811 pKa = 9.18EE812 pKa = 4.37GGSLTLNNDD821 pKa = 3.11SFEE824 pKa = 4.47NNQDD828 pKa = 3.54GLLGASDD835 pKa = 3.59TSGAITITNSVFSHH849 pKa = 6.39NGTGDD854 pKa = 3.34GHH856 pKa = 4.99THH858 pKa = 5.77NLYY861 pKa = 10.34VGEE864 pKa = 4.32VGTLTVTGSTLTDD877 pKa = 2.9AVVGHH882 pKa = 6.56EE883 pKa = 4.1IKK885 pKa = 10.76SRR887 pKa = 11.84ADD889 pKa = 3.15TTIIQGNTISDD900 pKa = 4.07GPTGTGSYY908 pKa = 10.59SIDD911 pKa = 3.56LPNGGKK917 pKa = 9.93AVISGNVIEE926 pKa = 5.11KK927 pKa = 10.5GPNAGNPAIISVGEE941 pKa = 3.97EE942 pKa = 3.7GGVYY946 pKa = 10.52ANSSVTLSNNTVLNDD961 pKa = 3.45NASPSVVLVKK971 pKa = 10.82NDD973 pKa = 3.42TTGPATISGTSVWNLTASEE992 pKa = 4.18MLQGPGSVSGTTLLASEE1009 pKa = 4.66PSIAAAAAPTPTPTPTPAPTPTPTPTPTPADD1040 pKa = 3.77TPIVVAPGGTLTTAAGTTNTVTMPTGDD1067 pKa = 3.33ATVVSGGTDD1076 pKa = 3.55TIYY1079 pKa = 11.04AGAGSDD1085 pKa = 3.95KK1086 pKa = 10.56ISSGVNTVTVNQGSGPLTFIGGYY1109 pKa = 9.55GGNATLNMAAGGTGANLTLYY1129 pKa = 11.14GNTTINAAGTFTANDD1144 pKa = 3.92GGSGAGADD1152 pKa = 3.55VFNITKK1158 pKa = 10.59GDD1160 pKa = 3.58VTNITITNFGDD1171 pKa = 3.56AFHH1174 pKa = 7.36LIGFSAGEE1182 pKa = 3.93AQQAVATAQHH1192 pKa = 5.99TGAGEE1197 pKa = 4.08VMSFSDD1203 pKa = 4.63GSQVTLAGWNHH1214 pKa = 6.31AMLSWFQQ1221 pKa = 3.24
Molecular weight: 123.6 kDa
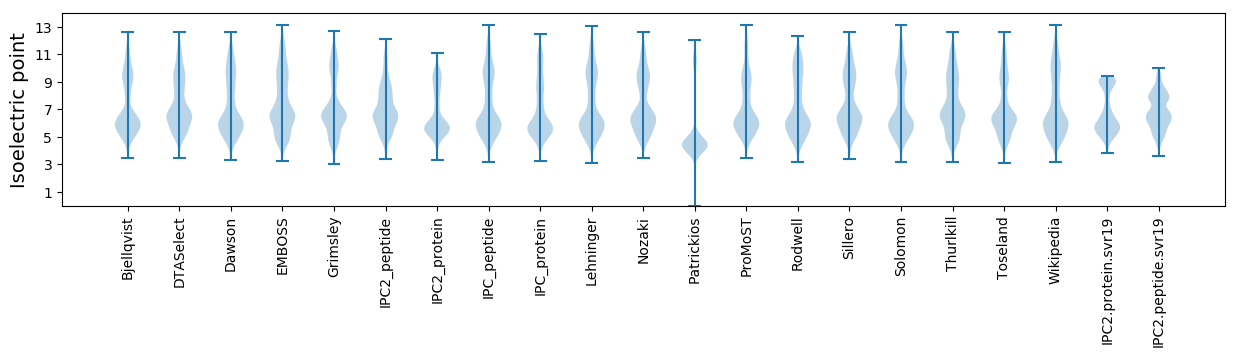
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D6P3Q0|A0A0D6P3Q0_9PROT Transcription termination factor Rho OS=Acidisphaera rubrifaciens HS-AP3 OX=1231350 GN=rho PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.09VLANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.24GRR39 pKa = 11.84QRR41 pKa = 11.84LSAA44 pKa = 4.04
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.09VLANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.24GRR39 pKa = 11.84QRR41 pKa = 11.84LSAA44 pKa = 4.04
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
851939 |
39 |
2198 |
305.6 |
32.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.238 ± 0.071 | 0.939 ± 0.016 |
5.824 ± 0.039 | 4.711 ± 0.05 |
3.349 ± 0.026 | 9.138 ± 0.051 |
2.316 ± 0.022 | 4.564 ± 0.031 |
1.978 ± 0.034 | 10.079 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.398 ± 0.022 | 2.173 ± 0.032 |
5.846 ± 0.038 | 2.807 ± 0.03 |
7.98 ± 0.064 | 4.499 ± 0.039 |
5.626 ± 0.05 | 7.777 ± 0.036 |
1.424 ± 0.018 | 2.334 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
