
Croceivirga radicis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Croceivirga
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
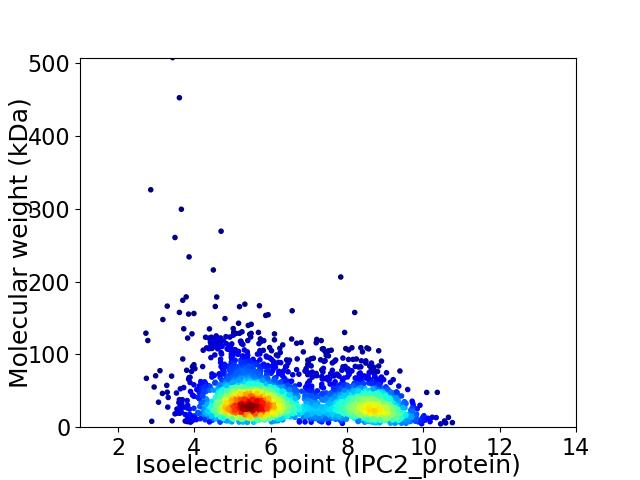
Virtual 2D-PAGE plot for 3064 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V6LPX4|A0A1V6LPX4_9FLAO SAM-dependent methyltransferase OS=Croceivirga radicis OX=1929488 GN=BUL40_12190 PE=4 SV=1
MM1 pKa = 7.14TKK3 pKa = 10.23FRR5 pKa = 11.84ICVIHH10 pKa = 6.99CIDD13 pKa = 3.62YY14 pKa = 10.87LRR16 pKa = 11.84RR17 pKa = 11.84TATFSYY23 pKa = 10.62LILAFVFSIQQLSAQTTFTEE43 pKa = 4.67SAASYY48 pKa = 10.19NLNIGQSKK56 pKa = 10.86DD57 pKa = 3.5GGHH60 pKa = 6.62AWADD64 pKa = 3.36FDD66 pKa = 6.27LDD68 pKa = 3.33GDD70 pKa = 3.83QDD72 pKa = 4.05VLVLVNSTSQKK83 pKa = 10.21ARR85 pKa = 11.84LMRR88 pKa = 11.84NNGDD92 pKa = 3.5GTFTNVQTTLAPGILNARR110 pKa = 11.84AEE112 pKa = 4.22RR113 pKa = 11.84QAAWGDD119 pKa = 3.68LNNDD123 pKa = 3.09GRR125 pKa = 11.84PDD127 pKa = 3.69FMITSSGNSGGNPSPPALQIFLQNLDD153 pKa = 3.69GTFGDD158 pKa = 4.68ALGGTNPITVGRR170 pKa = 11.84NGHH173 pKa = 6.55TITINPMNAEE183 pKa = 3.88GAGFFDD189 pKa = 5.29FEE191 pKa = 5.45GDD193 pKa = 3.18GDD195 pKa = 4.44LDD197 pKa = 4.12IFFDD201 pKa = 3.52SHH203 pKa = 7.59NYY205 pKa = 9.73GIEE208 pKa = 3.78ILRR211 pKa = 11.84NNYY214 pKa = 8.86IDD216 pKa = 3.84HH217 pKa = 6.74TNSTVTNPTAALLFSHH233 pKa = 6.64ITTGNGNGVVNFGLNQFATDD253 pKa = 3.59GDD255 pKa = 4.38YY256 pKa = 10.12GTAADD261 pKa = 4.69ANDD264 pKa = 5.01DD265 pKa = 3.6GWVDD269 pKa = 2.52IFMRR273 pKa = 11.84KK274 pKa = 8.61RR275 pKa = 11.84DD276 pKa = 3.74EE277 pKa = 4.48NDD279 pKa = 2.67FFLNQGGIFTNGSDD293 pKa = 4.22LAQAANGNKK302 pKa = 9.6GGNGLWDD309 pKa = 4.35LDD311 pKa = 3.81NDD313 pKa = 4.14GDD315 pKa = 5.06LDD317 pKa = 4.86AVWTEE322 pKa = 3.76NGLTQIYY329 pKa = 9.96RR330 pKa = 11.84NDD332 pKa = 3.5GSGIWTALGTGVFPGLPQPNNSNNGNSSNRR362 pKa = 11.84IDD364 pKa = 3.82ALAGGDD370 pKa = 3.72IDD372 pKa = 4.66NDD374 pKa = 3.56GDD376 pKa = 3.96IDD378 pKa = 3.8ILLVGQSRR386 pKa = 11.84SYY388 pKa = 11.44LYY390 pKa = 10.31INQLNSPTPAPGVIGSGSPMNFSLDD415 pKa = 3.34PEE417 pKa = 4.62TFNTNDD423 pKa = 3.64GEE425 pKa = 4.61GTTMVDD431 pKa = 3.05VDD433 pKa = 5.18DD434 pKa = 6.66DD435 pKa = 4.8GDD437 pKa = 4.09LDD439 pKa = 3.71IYY441 pKa = 11.01MNINGSGNQLWINNLAAANRR461 pKa = 11.84NNHH464 pKa = 5.71LMVNVIEE471 pKa = 5.01DD472 pKa = 3.62RR473 pKa = 11.84ASNGDD478 pKa = 3.16TGGFAGRR485 pKa = 11.84IALGTNVLIRR495 pKa = 11.84DD496 pKa = 3.73CSGNIVSGLRR506 pKa = 11.84QVNGVYY512 pKa = 10.51GHH514 pKa = 6.51GTQQPEE520 pKa = 4.33TVHH523 pKa = 6.64FGLPLGEE530 pKa = 4.15NEE532 pKa = 4.18TYY534 pKa = 10.13IIEE537 pKa = 4.04VRR539 pKa = 11.84YY540 pKa = 9.64PNFNDD545 pKa = 3.45PEE547 pKa = 4.11EE548 pKa = 5.28GITRR552 pKa = 11.84LIATGVAQPSTIAGTNHH569 pKa = 6.02YY570 pKa = 11.07NLTTTNAEE578 pKa = 4.32TIQNLNAPEE587 pKa = 5.24AEE589 pKa = 4.76DD590 pKa = 4.07DD591 pKa = 4.73LVTIPYY597 pKa = 9.09GNSVSIQINLFTNDD611 pKa = 3.92SEE613 pKa = 4.64PDD615 pKa = 3.43GEE617 pKa = 4.41NFFIEE622 pKa = 5.71SITQPTVGSVTIDD635 pKa = 3.35DD636 pKa = 4.53ANNGLVTYY644 pKa = 8.05TYY646 pKa = 11.3NNAAPFPGSTSFTYY660 pKa = 10.06TIVDD664 pKa = 3.42DD665 pKa = 4.54TYY667 pKa = 11.38NICAAQGKK675 pKa = 9.79SDD677 pKa = 3.6TATVTVYY684 pKa = 9.86EE685 pKa = 4.42PCSDD689 pKa = 3.58PTGVDD694 pKa = 3.01TDD696 pKa = 3.87GDD698 pKa = 4.69GINDD702 pKa = 3.23ICDD705 pKa = 3.47YY706 pKa = 11.71DD707 pKa = 4.59DD708 pKa = 5.9DD709 pKa = 5.38NDD711 pKa = 5.0GISDD715 pKa = 4.27FDD717 pKa = 3.83EE718 pKa = 4.56QQCSTTNPGVLGVPTTVLGGTAVDD742 pKa = 5.36EE743 pKa = 5.24IYY745 pKa = 10.77TDD747 pKa = 4.07YY748 pKa = 11.45NGYY751 pKa = 9.22WYY753 pKa = 10.87SSTASINSIEE763 pKa = 4.44PNLSHH768 pKa = 7.02NLLAFGSGGNLYY780 pKa = 9.14STGVVNTNMLDD791 pKa = 3.33TDD793 pKa = 4.0SNGRR797 pKa = 11.84YY798 pKa = 9.59DD799 pKa = 5.08AVDD802 pKa = 3.49TNGDD806 pKa = 2.98GSGNINITEE815 pKa = 4.16MEE817 pKa = 3.78WRR819 pKa = 11.84AFRR822 pKa = 11.84PNSNITNKK830 pKa = 9.75VALEE834 pKa = 3.97SALNDD839 pKa = 3.65GTLGSALGSTVIANLATDD857 pKa = 4.45PLNPLLTNGPRR868 pKa = 11.84GLDD871 pKa = 3.4LGTGIANVFDD881 pKa = 3.73SWFFNISTIDD891 pKa = 3.14IAAVNDD897 pKa = 4.21GLPDD901 pKa = 3.48ILITQVAQISSTVDD915 pKa = 2.84HH916 pKa = 6.96NIAFFDD922 pKa = 3.66AAGNPVGNQVRR933 pKa = 11.84VTSLGGGEE941 pKa = 4.13LSSVIGAQRR950 pKa = 11.84YY951 pKa = 8.4DD952 pKa = 3.35VYY954 pKa = 10.3QTNGSSWQVNVTKK967 pKa = 10.43NIRR970 pKa = 11.84MATLEE975 pKa = 4.04LSEE978 pKa = 5.04FGLDD982 pKa = 3.24ASNIGNVALMRR993 pKa = 11.84LTLNNSADD1001 pKa = 4.02TAFLAYY1007 pKa = 11.0NMDD1010 pKa = 3.83SFGEE1014 pKa = 4.3FCTDD1018 pKa = 4.17LDD1020 pKa = 3.97TDD1022 pKa = 3.65GDD1024 pKa = 4.59GIVDD1028 pKa = 4.55RR1029 pKa = 11.84LDD1031 pKa = 4.13LDD1033 pKa = 3.67SDD1035 pKa = 3.62NDD1037 pKa = 4.44GIYY1040 pKa = 10.89DD1041 pKa = 3.76VVEE1044 pKa = 4.41AGHH1047 pKa = 6.59NLTHH1051 pKa = 6.32TNGRR1055 pKa = 11.84LTGAVGADD1063 pKa = 4.07GIIDD1067 pKa = 4.09ALQTNPNNQTTTYY1080 pKa = 10.32IVADD1084 pKa = 4.0TNGDD1088 pKa = 4.14GIPDD1092 pKa = 4.45FKK1094 pKa = 11.52NLDD1097 pKa = 3.71SDD1099 pKa = 4.71GDD1101 pKa = 3.83GCTDD1105 pKa = 3.15VNEE1108 pKa = 5.32AGITDD1113 pKa = 3.95NNDD1116 pKa = 2.93DD1117 pKa = 4.73GYY1119 pKa = 11.52LGDD1122 pKa = 4.12IPLTVSLDD1130 pKa = 3.65GVVTSGADD1138 pKa = 3.6GYY1140 pKa = 6.54TTPVDD1145 pKa = 3.46ADD1147 pKa = 3.57GNGIYY1152 pKa = 10.19DD1153 pKa = 3.79YY1154 pKa = 10.27TEE1156 pKa = 3.82VGAAPTITDD1165 pKa = 3.34QPTNTTICPGCSGTITANFANADD1188 pKa = 3.71GYY1190 pKa = 10.44QWQLFNGSTWEE1201 pKa = 3.85NLVNGGLYY1209 pKa = 10.7GGTTTQTLTITNPTSAEE1226 pKa = 3.8HH1227 pKa = 6.34GNTYY1231 pKa = 10.3RR1232 pKa = 11.84IVGSSTTFVCRR1243 pKa = 11.84NAISNPVVLSVRR1255 pKa = 11.84INTVITNRR1263 pKa = 11.84RR1264 pKa = 11.84ITYY1267 pKa = 10.09RR1268 pKa = 11.84VDD1270 pKa = 3.17PKK1272 pKa = 11.36
MM1 pKa = 7.14TKK3 pKa = 10.23FRR5 pKa = 11.84ICVIHH10 pKa = 6.99CIDD13 pKa = 3.62YY14 pKa = 10.87LRR16 pKa = 11.84RR17 pKa = 11.84TATFSYY23 pKa = 10.62LILAFVFSIQQLSAQTTFTEE43 pKa = 4.67SAASYY48 pKa = 10.19NLNIGQSKK56 pKa = 10.86DD57 pKa = 3.5GGHH60 pKa = 6.62AWADD64 pKa = 3.36FDD66 pKa = 6.27LDD68 pKa = 3.33GDD70 pKa = 3.83QDD72 pKa = 4.05VLVLVNSTSQKK83 pKa = 10.21ARR85 pKa = 11.84LMRR88 pKa = 11.84NNGDD92 pKa = 3.5GTFTNVQTTLAPGILNARR110 pKa = 11.84AEE112 pKa = 4.22RR113 pKa = 11.84QAAWGDD119 pKa = 3.68LNNDD123 pKa = 3.09GRR125 pKa = 11.84PDD127 pKa = 3.69FMITSSGNSGGNPSPPALQIFLQNLDD153 pKa = 3.69GTFGDD158 pKa = 4.68ALGGTNPITVGRR170 pKa = 11.84NGHH173 pKa = 6.55TITINPMNAEE183 pKa = 3.88GAGFFDD189 pKa = 5.29FEE191 pKa = 5.45GDD193 pKa = 3.18GDD195 pKa = 4.44LDD197 pKa = 4.12IFFDD201 pKa = 3.52SHH203 pKa = 7.59NYY205 pKa = 9.73GIEE208 pKa = 3.78ILRR211 pKa = 11.84NNYY214 pKa = 8.86IDD216 pKa = 3.84HH217 pKa = 6.74TNSTVTNPTAALLFSHH233 pKa = 6.64ITTGNGNGVVNFGLNQFATDD253 pKa = 3.59GDD255 pKa = 4.38YY256 pKa = 10.12GTAADD261 pKa = 4.69ANDD264 pKa = 5.01DD265 pKa = 3.6GWVDD269 pKa = 2.52IFMRR273 pKa = 11.84KK274 pKa = 8.61RR275 pKa = 11.84DD276 pKa = 3.74EE277 pKa = 4.48NDD279 pKa = 2.67FFLNQGGIFTNGSDD293 pKa = 4.22LAQAANGNKK302 pKa = 9.6GGNGLWDD309 pKa = 4.35LDD311 pKa = 3.81NDD313 pKa = 4.14GDD315 pKa = 5.06LDD317 pKa = 4.86AVWTEE322 pKa = 3.76NGLTQIYY329 pKa = 9.96RR330 pKa = 11.84NDD332 pKa = 3.5GSGIWTALGTGVFPGLPQPNNSNNGNSSNRR362 pKa = 11.84IDD364 pKa = 3.82ALAGGDD370 pKa = 3.72IDD372 pKa = 4.66NDD374 pKa = 3.56GDD376 pKa = 3.96IDD378 pKa = 3.8ILLVGQSRR386 pKa = 11.84SYY388 pKa = 11.44LYY390 pKa = 10.31INQLNSPTPAPGVIGSGSPMNFSLDD415 pKa = 3.34PEE417 pKa = 4.62TFNTNDD423 pKa = 3.64GEE425 pKa = 4.61GTTMVDD431 pKa = 3.05VDD433 pKa = 5.18DD434 pKa = 6.66DD435 pKa = 4.8GDD437 pKa = 4.09LDD439 pKa = 3.71IYY441 pKa = 11.01MNINGSGNQLWINNLAAANRR461 pKa = 11.84NNHH464 pKa = 5.71LMVNVIEE471 pKa = 5.01DD472 pKa = 3.62RR473 pKa = 11.84ASNGDD478 pKa = 3.16TGGFAGRR485 pKa = 11.84IALGTNVLIRR495 pKa = 11.84DD496 pKa = 3.73CSGNIVSGLRR506 pKa = 11.84QVNGVYY512 pKa = 10.51GHH514 pKa = 6.51GTQQPEE520 pKa = 4.33TVHH523 pKa = 6.64FGLPLGEE530 pKa = 4.15NEE532 pKa = 4.18TYY534 pKa = 10.13IIEE537 pKa = 4.04VRR539 pKa = 11.84YY540 pKa = 9.64PNFNDD545 pKa = 3.45PEE547 pKa = 4.11EE548 pKa = 5.28GITRR552 pKa = 11.84LIATGVAQPSTIAGTNHH569 pKa = 6.02YY570 pKa = 11.07NLTTTNAEE578 pKa = 4.32TIQNLNAPEE587 pKa = 5.24AEE589 pKa = 4.76DD590 pKa = 4.07DD591 pKa = 4.73LVTIPYY597 pKa = 9.09GNSVSIQINLFTNDD611 pKa = 3.92SEE613 pKa = 4.64PDD615 pKa = 3.43GEE617 pKa = 4.41NFFIEE622 pKa = 5.71SITQPTVGSVTIDD635 pKa = 3.35DD636 pKa = 4.53ANNGLVTYY644 pKa = 8.05TYY646 pKa = 11.3NNAAPFPGSTSFTYY660 pKa = 10.06TIVDD664 pKa = 3.42DD665 pKa = 4.54TYY667 pKa = 11.38NICAAQGKK675 pKa = 9.79SDD677 pKa = 3.6TATVTVYY684 pKa = 9.86EE685 pKa = 4.42PCSDD689 pKa = 3.58PTGVDD694 pKa = 3.01TDD696 pKa = 3.87GDD698 pKa = 4.69GINDD702 pKa = 3.23ICDD705 pKa = 3.47YY706 pKa = 11.71DD707 pKa = 4.59DD708 pKa = 5.9DD709 pKa = 5.38NDD711 pKa = 5.0GISDD715 pKa = 4.27FDD717 pKa = 3.83EE718 pKa = 4.56QQCSTTNPGVLGVPTTVLGGTAVDD742 pKa = 5.36EE743 pKa = 5.24IYY745 pKa = 10.77TDD747 pKa = 4.07YY748 pKa = 11.45NGYY751 pKa = 9.22WYY753 pKa = 10.87SSTASINSIEE763 pKa = 4.44PNLSHH768 pKa = 7.02NLLAFGSGGNLYY780 pKa = 9.14STGVVNTNMLDD791 pKa = 3.33TDD793 pKa = 4.0SNGRR797 pKa = 11.84YY798 pKa = 9.59DD799 pKa = 5.08AVDD802 pKa = 3.49TNGDD806 pKa = 2.98GSGNINITEE815 pKa = 4.16MEE817 pKa = 3.78WRR819 pKa = 11.84AFRR822 pKa = 11.84PNSNITNKK830 pKa = 9.75VALEE834 pKa = 3.97SALNDD839 pKa = 3.65GTLGSALGSTVIANLATDD857 pKa = 4.45PLNPLLTNGPRR868 pKa = 11.84GLDD871 pKa = 3.4LGTGIANVFDD881 pKa = 3.73SWFFNISTIDD891 pKa = 3.14IAAVNDD897 pKa = 4.21GLPDD901 pKa = 3.48ILITQVAQISSTVDD915 pKa = 2.84HH916 pKa = 6.96NIAFFDD922 pKa = 3.66AAGNPVGNQVRR933 pKa = 11.84VTSLGGGEE941 pKa = 4.13LSSVIGAQRR950 pKa = 11.84YY951 pKa = 8.4DD952 pKa = 3.35VYY954 pKa = 10.3QTNGSSWQVNVTKK967 pKa = 10.43NIRR970 pKa = 11.84MATLEE975 pKa = 4.04LSEE978 pKa = 5.04FGLDD982 pKa = 3.24ASNIGNVALMRR993 pKa = 11.84LTLNNSADD1001 pKa = 4.02TAFLAYY1007 pKa = 11.0NMDD1010 pKa = 3.83SFGEE1014 pKa = 4.3FCTDD1018 pKa = 4.17LDD1020 pKa = 3.97TDD1022 pKa = 3.65GDD1024 pKa = 4.59GIVDD1028 pKa = 4.55RR1029 pKa = 11.84LDD1031 pKa = 4.13LDD1033 pKa = 3.67SDD1035 pKa = 3.62NDD1037 pKa = 4.44GIYY1040 pKa = 10.89DD1041 pKa = 3.76VVEE1044 pKa = 4.41AGHH1047 pKa = 6.59NLTHH1051 pKa = 6.32TNGRR1055 pKa = 11.84LTGAVGADD1063 pKa = 4.07GIIDD1067 pKa = 4.09ALQTNPNNQTTTYY1080 pKa = 10.32IVADD1084 pKa = 4.0TNGDD1088 pKa = 4.14GIPDD1092 pKa = 4.45FKK1094 pKa = 11.52NLDD1097 pKa = 3.71SDD1099 pKa = 4.71GDD1101 pKa = 3.83GCTDD1105 pKa = 3.15VNEE1108 pKa = 5.32AGITDD1113 pKa = 3.95NNDD1116 pKa = 2.93DD1117 pKa = 4.73GYY1119 pKa = 11.52LGDD1122 pKa = 4.12IPLTVSLDD1130 pKa = 3.65GVVTSGADD1138 pKa = 3.6GYY1140 pKa = 6.54TTPVDD1145 pKa = 3.46ADD1147 pKa = 3.57GNGIYY1152 pKa = 10.19DD1153 pKa = 3.79YY1154 pKa = 10.27TEE1156 pKa = 3.82VGAAPTITDD1165 pKa = 3.34QPTNTTICPGCSGTITANFANADD1188 pKa = 3.71GYY1190 pKa = 10.44QWQLFNGSTWEE1201 pKa = 3.85NLVNGGLYY1209 pKa = 10.7GGTTTQTLTITNPTSAEE1226 pKa = 3.8HH1227 pKa = 6.34GNTYY1231 pKa = 10.3RR1232 pKa = 11.84IVGSSTTFVCRR1243 pKa = 11.84NAISNPVVLSVRR1255 pKa = 11.84INTVITNRR1263 pKa = 11.84RR1264 pKa = 11.84ITYY1267 pKa = 10.09RR1268 pKa = 11.84VDD1270 pKa = 3.17PKK1272 pKa = 11.36
Molecular weight: 135.2 kDa
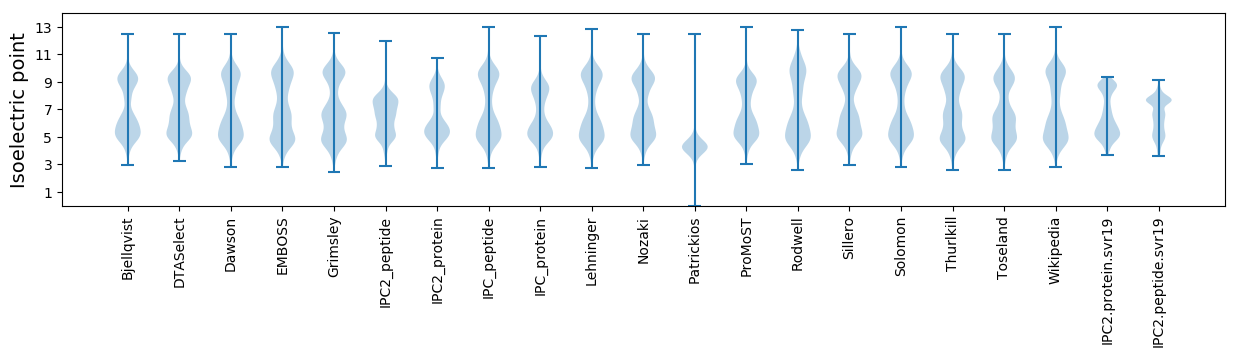
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V6LTL2|A0A1V6LTL2_9FLAO Bifunctional alpha alpha-trehalose-phosphate synthase (UDP-forming)/trehalose-phosphatase OS=Croceivirga radicis OX=1929488 GN=BUL40_06645 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.19RR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.08GRR39 pKa = 11.84KK40 pKa = 8.3KK41 pKa = 9.83LTVSSEE47 pKa = 3.9PRR49 pKa = 11.84HH50 pKa = 5.77KK51 pKa = 10.61KK52 pKa = 9.84
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.19RR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.08GRR39 pKa = 11.84KK40 pKa = 8.3KK41 pKa = 9.83LTVSSEE47 pKa = 3.9PRR49 pKa = 11.84HH50 pKa = 5.77KK51 pKa = 10.61KK52 pKa = 9.84
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1106336 |
38 |
4776 |
361.1 |
40.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.085 ± 0.045 | 0.704 ± 0.017 |
5.619 ± 0.044 | 6.501 ± 0.041 |
5.073 ± 0.03 | 6.816 ± 0.052 |
1.75 ± 0.024 | 7.09 ± 0.041 |
7.355 ± 0.073 | 9.654 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.132 ± 0.022 | 6.051 ± 0.04 |
3.604 ± 0.029 | 3.582 ± 0.025 |
3.398 ± 0.028 | 5.868 ± 0.031 |
6.093 ± 0.064 | 6.422 ± 0.033 |
1.138 ± 0.016 | 4.066 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
