
Strongyloides stercoralis (Threadworm)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Tylenchina; Panagrolaimomorpha; Strongyloidoidea; Strongyloididae; Strongyloides
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
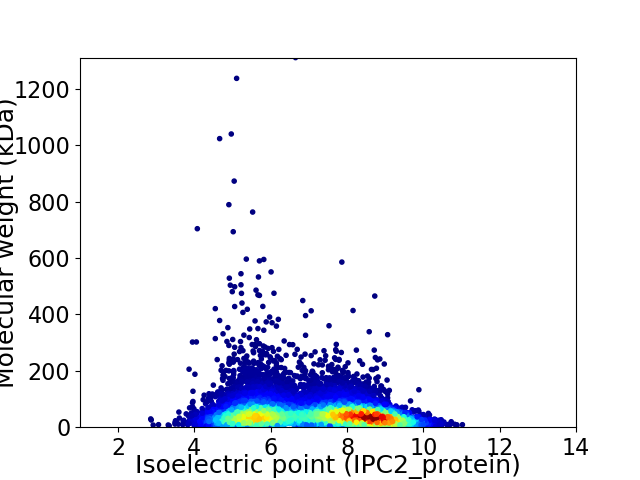
Virtual 2D-PAGE plot for 12851 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K0DVY7|A0A0K0DVY7_STRER Uncharacterized protein OS=Strongyloides stercoralis OX=6248 PE=4 SV=1
MM1 pKa = 7.25ITLQEE6 pKa = 3.78VSIPRR11 pKa = 11.84EE12 pKa = 4.21GVCFTEE18 pKa = 4.51EE19 pKa = 3.83NVQLFHH25 pKa = 6.9EE26 pKa = 4.61VEE28 pKa = 4.3KK29 pKa = 11.21VSNGTLYY36 pKa = 10.28IGEE39 pKa = 4.6RR40 pKa = 11.84NITWIEE46 pKa = 3.94DD47 pKa = 3.5GKK49 pKa = 11.22GLGIIIPYY57 pKa = 8.61QALVTHH63 pKa = 7.15GISNDD68 pKa = 2.59INAFPDD74 pKa = 2.99RR75 pKa = 11.84CLFLTIDD82 pKa = 3.39ASKK85 pKa = 11.52VNFNVSSSEE94 pKa = 3.65EE95 pKa = 4.18DD96 pKa = 2.98MDD98 pKa = 5.56NDD100 pKa = 5.03DD101 pKa = 4.83EE102 pKa = 4.75YY103 pKa = 11.49EE104 pKa = 4.41CIYY107 pKa = 10.63LRR109 pKa = 11.84LVPANDD115 pKa = 3.55SLNIMFQMMTEE126 pKa = 4.07CQEE129 pKa = 4.07MNPDD133 pKa = 3.79EE134 pKa = 4.88EE135 pKa = 5.51DD136 pKa = 3.43MEE138 pKa = 4.85EE139 pKa = 4.46DD140 pKa = 3.9FSSGLQVVNGGSSDD154 pKa = 3.21WYY156 pKa = 10.45GVTDD160 pKa = 4.2NEE162 pKa = 4.73PFPDD166 pKa = 3.68MSDD169 pKa = 2.94QGMANYY175 pKa = 10.02DD176 pKa = 3.39RR177 pKa = 11.84LFGNKK182 pKa = 9.56NNDD185 pKa = 3.32SMDD188 pKa = 3.82DD189 pKa = 3.32
MM1 pKa = 7.25ITLQEE6 pKa = 3.78VSIPRR11 pKa = 11.84EE12 pKa = 4.21GVCFTEE18 pKa = 4.51EE19 pKa = 3.83NVQLFHH25 pKa = 6.9EE26 pKa = 4.61VEE28 pKa = 4.3KK29 pKa = 11.21VSNGTLYY36 pKa = 10.28IGEE39 pKa = 4.6RR40 pKa = 11.84NITWIEE46 pKa = 3.94DD47 pKa = 3.5GKK49 pKa = 11.22GLGIIIPYY57 pKa = 8.61QALVTHH63 pKa = 7.15GISNDD68 pKa = 2.59INAFPDD74 pKa = 2.99RR75 pKa = 11.84CLFLTIDD82 pKa = 3.39ASKK85 pKa = 11.52VNFNVSSSEE94 pKa = 3.65EE95 pKa = 4.18DD96 pKa = 2.98MDD98 pKa = 5.56NDD100 pKa = 5.03DD101 pKa = 4.83EE102 pKa = 4.75YY103 pKa = 11.49EE104 pKa = 4.41CIYY107 pKa = 10.63LRR109 pKa = 11.84LVPANDD115 pKa = 3.55SLNIMFQMMTEE126 pKa = 4.07CQEE129 pKa = 4.07MNPDD133 pKa = 3.79EE134 pKa = 4.88EE135 pKa = 5.51DD136 pKa = 3.43MEE138 pKa = 4.85EE139 pKa = 4.46DD140 pKa = 3.9FSSGLQVVNGGSSDD154 pKa = 3.21WYY156 pKa = 10.45GVTDD160 pKa = 4.2NEE162 pKa = 4.73PFPDD166 pKa = 3.68MSDD169 pKa = 2.94QGMANYY175 pKa = 10.02DD176 pKa = 3.39RR177 pKa = 11.84LFGNKK182 pKa = 9.56NNDD185 pKa = 3.32SMDD188 pKa = 3.82DD189 pKa = 3.32
Molecular weight: 21.4 kDa
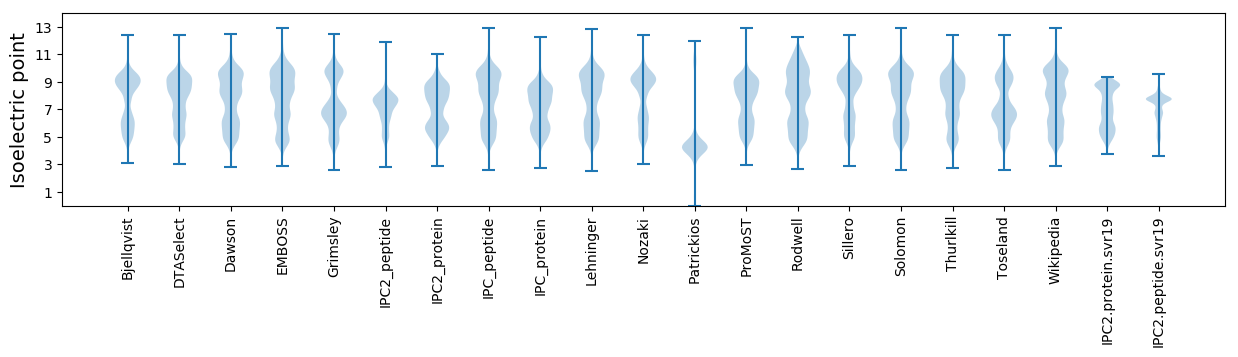
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K0DUM0|A0A0K0DUM0_STRER Uncharacterized protein OS=Strongyloides stercoralis OX=6248 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.54FIFSSVLVIILLSHH16 pKa = 7.38KK17 pKa = 10.23IFSQQLSVQPAQTKK31 pKa = 9.97IFAAPVFTSNNAIEE45 pKa = 4.33IDD47 pKa = 3.96GEE49 pKa = 4.24KK50 pKa = 10.37KK51 pKa = 10.32RR52 pKa = 11.84LFKK55 pKa = 10.7IGAARR60 pKa = 11.84DD61 pKa = 4.03GVQKK65 pKa = 10.97AILDD69 pKa = 4.01EE70 pKa = 4.34VQPSQTVDD78 pKa = 3.92DD79 pKa = 4.35ISDD82 pKa = 3.69FNEE85 pKa = 4.06DD86 pKa = 2.96NTIFGAYY93 pKa = 9.68DD94 pKa = 3.46EE95 pKa = 5.49EE96 pKa = 4.37IVEE99 pKa = 4.47PVKK102 pKa = 9.63EE103 pKa = 4.31TPIFIPKK110 pKa = 10.16LYY112 pKa = 10.29RR113 pKa = 11.84ADD115 pKa = 3.59KK116 pKa = 10.87DD117 pKa = 3.77EE118 pKa = 4.47PTTTTTTTTTTTTRR132 pKa = 11.84RR133 pKa = 11.84PTTTRR138 pKa = 11.84QTVTTRR144 pKa = 11.84ITTLAPHH151 pKa = 5.23TTVRR155 pKa = 11.84TIQNNRR161 pKa = 11.84IPQTNQINSIPQSFVPSRR179 pKa = 11.84MISPPRR185 pKa = 11.84QPQQISGLQTIPPQIVPRR203 pKa = 11.84PNVIQPLPQQQFQFRR218 pKa = 11.84QNIRR222 pKa = 11.84PQPVLQTRR230 pKa = 11.84TGFSTPPPRR239 pKa = 11.84VISTTLRR246 pKa = 11.84PIITRR251 pKa = 11.84QPQTFPTLAPVTSTSRR267 pKa = 11.84PLRR270 pKa = 11.84PFIPQQPNPRR280 pKa = 11.84NSQNLRR286 pKa = 11.84TGVCRR291 pKa = 11.84ASIFYY296 pKa = 8.26LTSVPSSQDD305 pKa = 2.9FQTQFTHH312 pKa = 6.09FAVVVSVDD320 pKa = 3.11QCARR324 pKa = 11.84TCHH327 pKa = 5.73EE328 pKa = 4.67FNCAVAIYY336 pKa = 9.85DD337 pKa = 4.33ASTKK341 pKa = 9.93HH342 pKa = 5.86CQFVPSTAFSIRR354 pKa = 11.84EE355 pKa = 4.18GQCPNWPNPLYY366 pKa = 10.61RR367 pKa = 11.84NNVLGLNNIRR377 pKa = 11.84ISCVTCQRR385 pKa = 11.84RR386 pKa = 11.84RR387 pKa = 11.84RR388 pKa = 11.84NFQRR392 pKa = 11.84VRR394 pKa = 11.84GMSRR398 pKa = 11.84GFGLTNRR405 pKa = 11.84RR406 pKa = 11.84QPTTIQTTRR415 pKa = 11.84FLPQTPNRR423 pKa = 11.84NIPTNPQNSNIRR435 pKa = 11.84LNGVILKK442 pKa = 10.09QPTSYY447 pKa = 10.79AISSNEE453 pKa = 3.7MEE455 pKa = 4.58KK456 pKa = 10.88LLSEE460 pKa = 4.2SSITKK465 pKa = 10.2GSQLSEE471 pKa = 3.9INAA474 pKa = 3.86
MM1 pKa = 7.44KK2 pKa = 10.54FIFSSVLVIILLSHH16 pKa = 7.38KK17 pKa = 10.23IFSQQLSVQPAQTKK31 pKa = 9.97IFAAPVFTSNNAIEE45 pKa = 4.33IDD47 pKa = 3.96GEE49 pKa = 4.24KK50 pKa = 10.37KK51 pKa = 10.32RR52 pKa = 11.84LFKK55 pKa = 10.7IGAARR60 pKa = 11.84DD61 pKa = 4.03GVQKK65 pKa = 10.97AILDD69 pKa = 4.01EE70 pKa = 4.34VQPSQTVDD78 pKa = 3.92DD79 pKa = 4.35ISDD82 pKa = 3.69FNEE85 pKa = 4.06DD86 pKa = 2.96NTIFGAYY93 pKa = 9.68DD94 pKa = 3.46EE95 pKa = 5.49EE96 pKa = 4.37IVEE99 pKa = 4.47PVKK102 pKa = 9.63EE103 pKa = 4.31TPIFIPKK110 pKa = 10.16LYY112 pKa = 10.29RR113 pKa = 11.84ADD115 pKa = 3.59KK116 pKa = 10.87DD117 pKa = 3.77EE118 pKa = 4.47PTTTTTTTTTTTTRR132 pKa = 11.84RR133 pKa = 11.84PTTTRR138 pKa = 11.84QTVTTRR144 pKa = 11.84ITTLAPHH151 pKa = 5.23TTVRR155 pKa = 11.84TIQNNRR161 pKa = 11.84IPQTNQINSIPQSFVPSRR179 pKa = 11.84MISPPRR185 pKa = 11.84QPQQISGLQTIPPQIVPRR203 pKa = 11.84PNVIQPLPQQQFQFRR218 pKa = 11.84QNIRR222 pKa = 11.84PQPVLQTRR230 pKa = 11.84TGFSTPPPRR239 pKa = 11.84VISTTLRR246 pKa = 11.84PIITRR251 pKa = 11.84QPQTFPTLAPVTSTSRR267 pKa = 11.84PLRR270 pKa = 11.84PFIPQQPNPRR280 pKa = 11.84NSQNLRR286 pKa = 11.84TGVCRR291 pKa = 11.84ASIFYY296 pKa = 8.26LTSVPSSQDD305 pKa = 2.9FQTQFTHH312 pKa = 6.09FAVVVSVDD320 pKa = 3.11QCARR324 pKa = 11.84TCHH327 pKa = 5.73EE328 pKa = 4.67FNCAVAIYY336 pKa = 9.85DD337 pKa = 4.33ASTKK341 pKa = 9.93HH342 pKa = 5.86CQFVPSTAFSIRR354 pKa = 11.84EE355 pKa = 4.18GQCPNWPNPLYY366 pKa = 10.61RR367 pKa = 11.84NNVLGLNNIRR377 pKa = 11.84ISCVTCQRR385 pKa = 11.84RR386 pKa = 11.84RR387 pKa = 11.84RR388 pKa = 11.84NFQRR392 pKa = 11.84VRR394 pKa = 11.84GMSRR398 pKa = 11.84GFGLTNRR405 pKa = 11.84RR406 pKa = 11.84QPTTIQTTRR415 pKa = 11.84FLPQTPNRR423 pKa = 11.84NIPTNPQNSNIRR435 pKa = 11.84LNGVILKK442 pKa = 10.09QPTSYY447 pKa = 10.79AISSNEE453 pKa = 3.7MEE455 pKa = 4.58KK456 pKa = 10.88LLSEE460 pKa = 4.2SSITKK465 pKa = 10.2GSQLSEE471 pKa = 3.9INAA474 pKa = 3.86
Molecular weight: 53.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5840225 |
8 |
11712 |
454.5 |
52.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.884 ± 0.022 | 2.068 ± 0.019 |
5.441 ± 0.02 | 6.628 ± 0.03 |
4.914 ± 0.019 | 4.68 ± 0.025 |
1.948 ± 0.01 | 8.541 ± 0.022 |
8.703 ± 0.026 | 8.663 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.166 ± 0.01 | 7.677 ± 0.033 |
3.827 ± 0.017 | 3.351 ± 0.015 |
3.819 ± 0.018 | 8.02 ± 0.029 |
5.602 ± 0.03 | 5.07 ± 0.017 |
0.894 ± 0.007 | 4.101 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
