
Wenzhou tombus-like virus 18
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.22
Get precalculated fractions of proteins
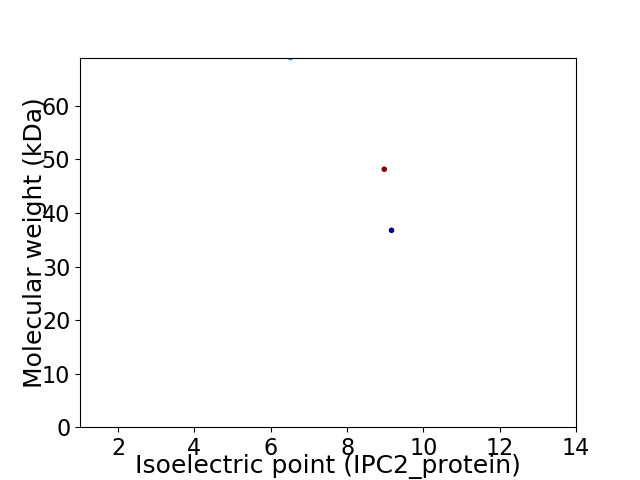
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
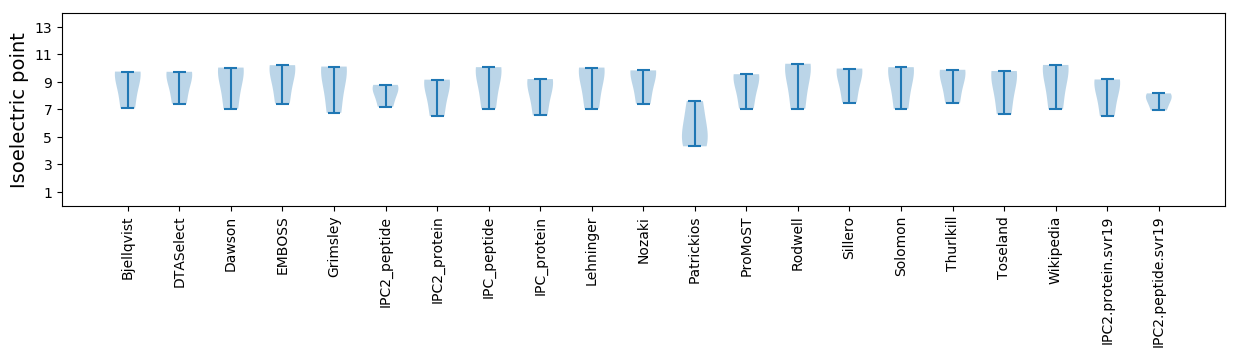
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFE3|A0A1L3KFE3_9VIRU RNA-directed RNA polymerase OS=Wenzhou tombus-like virus 18 OX=1923671 PE=4 SV=1
MM1 pKa = 7.7RR2 pKa = 11.84SIGTRR7 pKa = 11.84CVPSWLTHH15 pKa = 5.31KK16 pKa = 9.21TDD18 pKa = 4.01SGSSSPFEE26 pKa = 3.93TQGRR30 pKa = 11.84HH31 pKa = 5.57LLPPLLDD38 pKa = 3.85NLAPLPALTRR48 pKa = 11.84DD49 pKa = 3.44NQLAGIEE56 pKa = 4.15KK57 pKa = 10.0RR58 pKa = 11.84VLAVVPKK65 pKa = 8.49PHH67 pKa = 7.65PDD69 pKa = 3.26LVMEE73 pKa = 4.9DD74 pKa = 3.62QPKK77 pKa = 9.7PRR79 pKa = 11.84PFHH82 pKa = 6.46HH83 pKa = 7.04CPSVPTPEE91 pKa = 4.02RR92 pKa = 11.84SLIKK96 pKa = 10.77QMGDD100 pKa = 3.11LVASTGKK107 pKa = 9.86IPPVAAQPFLGIPPIYY123 pKa = 10.05LVPLSLMPDD132 pKa = 3.15DD133 pKa = 3.79WVEE136 pKa = 3.91EE137 pKa = 4.22VVDD140 pKa = 3.31IARR143 pKa = 11.84RR144 pKa = 11.84RR145 pKa = 11.84KK146 pKa = 9.52VQDD149 pKa = 3.88PMLPWPTAPTSADD162 pKa = 3.23LHH164 pKa = 6.76DD165 pKa = 4.99SSSCLLGPVTCRR177 pKa = 11.84TDD179 pKa = 3.36RR180 pKa = 11.84LNHH183 pKa = 5.93PFFRR187 pKa = 11.84CNLTSLVHH195 pKa = 6.0EE196 pKa = 5.04FWKK199 pKa = 10.63LRR201 pKa = 11.84EE202 pKa = 4.24NQWLTALEE210 pKa = 4.12PLPFEE215 pKa = 4.18EE216 pKa = 4.24WLEE219 pKa = 3.98RR220 pKa = 11.84FPNARR225 pKa = 11.84RR226 pKa = 11.84QQLRR230 pKa = 11.84DD231 pKa = 3.49ARR233 pKa = 11.84EE234 pKa = 3.3EE235 pKa = 4.1AYY237 pKa = 10.6RR238 pKa = 11.84HH239 pKa = 5.13GLEE242 pKa = 4.19SKK244 pKa = 10.25DD245 pKa = 3.62AIMAVFIKK253 pKa = 10.06TEE255 pKa = 4.07TSTSGTDD262 pKa = 3.28PRR264 pKa = 11.84NISPRR269 pKa = 11.84QLKK272 pKa = 9.59FLSVLGPFVAAIEE285 pKa = 4.27KK286 pKa = 9.59QAKK289 pKa = 8.74CCDD292 pKa = 3.66YY293 pKa = 10.87LVKK296 pKa = 10.73GKK298 pKa = 10.08NPEE301 pKa = 3.66EE302 pKa = 3.92RR303 pKa = 11.84GQYY306 pKa = 9.1ISQYY310 pKa = 7.69WRR312 pKa = 11.84HH313 pKa = 5.58SVIEE317 pKa = 4.12TDD319 pKa = 4.58FSRR322 pKa = 11.84FDD324 pKa = 3.47MTVSADD330 pKa = 3.49FVKK333 pKa = 9.86TVEE336 pKa = 3.65RR337 pKa = 11.84SMFRR341 pKa = 11.84RR342 pKa = 11.84AFPEE346 pKa = 3.94GLYY349 pKa = 10.0PALDD353 pKa = 3.68EE354 pKa = 5.3CLDD357 pKa = 4.33LLPVMTGASSLGVLYY372 pKa = 10.89SVAGTRR378 pKa = 11.84ASGDD382 pKa = 3.08AHH384 pKa = 5.82TSIANGVLNRR394 pKa = 11.84FVIWASLRR402 pKa = 11.84NMDD405 pKa = 3.78PSTWVSFHH413 pKa = 6.84EE414 pKa = 4.71GDD416 pKa = 5.21DD417 pKa = 4.75GIIHH421 pKa = 7.24CDD423 pKa = 3.22SAVTQQICDD432 pKa = 3.78YY433 pKa = 10.4LAFAAFLGFKK443 pKa = 9.99LKK445 pKa = 10.28VVRR448 pKa = 11.84PPSHH452 pKa = 5.84EE453 pKa = 3.8HH454 pKa = 6.59ANFCGRR460 pKa = 11.84HH461 pKa = 4.15TCEE464 pKa = 3.42EE465 pKa = 4.29CHH467 pKa = 7.33RR468 pKa = 11.84EE469 pKa = 4.0FCDD472 pKa = 4.78LPRR475 pKa = 11.84ALSKK479 pKa = 9.62FHH481 pKa = 6.36ITFKK485 pKa = 10.37QGEE488 pKa = 4.3PRR490 pKa = 11.84SLLLAKK496 pKa = 10.2ALSYY500 pKa = 10.4FATDD504 pKa = 2.42AHH506 pKa = 6.03TPIIGTMCDD515 pKa = 2.84ALIRR519 pKa = 11.84HH520 pKa = 6.9LSPQVSDD527 pKa = 2.79RR528 pKa = 11.84LMRR531 pKa = 11.84RR532 pKa = 11.84RR533 pKa = 11.84LHH535 pKa = 6.67AMSRR539 pKa = 11.84YY540 pKa = 10.05DD541 pKa = 3.29RR542 pKa = 11.84DD543 pKa = 3.19RR544 pKa = 11.84TIRR547 pKa = 11.84GMHH550 pKa = 5.66NTFMSPSACCRR561 pKa = 11.84AAISNVCGWDD571 pKa = 3.37PKK573 pKa = 11.14LQVAWEE579 pKa = 4.06AQLATWRR586 pKa = 11.84YY587 pKa = 8.88GITEE591 pKa = 4.34ISPLSVEE598 pKa = 4.24DD599 pKa = 4.26HH600 pKa = 6.94LVDD603 pKa = 3.64TDD605 pKa = 4.36SVTIYY610 pKa = 10.5QGG612 pKa = 2.97
MM1 pKa = 7.7RR2 pKa = 11.84SIGTRR7 pKa = 11.84CVPSWLTHH15 pKa = 5.31KK16 pKa = 9.21TDD18 pKa = 4.01SGSSSPFEE26 pKa = 3.93TQGRR30 pKa = 11.84HH31 pKa = 5.57LLPPLLDD38 pKa = 3.85NLAPLPALTRR48 pKa = 11.84DD49 pKa = 3.44NQLAGIEE56 pKa = 4.15KK57 pKa = 10.0RR58 pKa = 11.84VLAVVPKK65 pKa = 8.49PHH67 pKa = 7.65PDD69 pKa = 3.26LVMEE73 pKa = 4.9DD74 pKa = 3.62QPKK77 pKa = 9.7PRR79 pKa = 11.84PFHH82 pKa = 6.46HH83 pKa = 7.04CPSVPTPEE91 pKa = 4.02RR92 pKa = 11.84SLIKK96 pKa = 10.77QMGDD100 pKa = 3.11LVASTGKK107 pKa = 9.86IPPVAAQPFLGIPPIYY123 pKa = 10.05LVPLSLMPDD132 pKa = 3.15DD133 pKa = 3.79WVEE136 pKa = 3.91EE137 pKa = 4.22VVDD140 pKa = 3.31IARR143 pKa = 11.84RR144 pKa = 11.84RR145 pKa = 11.84KK146 pKa = 9.52VQDD149 pKa = 3.88PMLPWPTAPTSADD162 pKa = 3.23LHH164 pKa = 6.76DD165 pKa = 4.99SSSCLLGPVTCRR177 pKa = 11.84TDD179 pKa = 3.36RR180 pKa = 11.84LNHH183 pKa = 5.93PFFRR187 pKa = 11.84CNLTSLVHH195 pKa = 6.0EE196 pKa = 5.04FWKK199 pKa = 10.63LRR201 pKa = 11.84EE202 pKa = 4.24NQWLTALEE210 pKa = 4.12PLPFEE215 pKa = 4.18EE216 pKa = 4.24WLEE219 pKa = 3.98RR220 pKa = 11.84FPNARR225 pKa = 11.84RR226 pKa = 11.84QQLRR230 pKa = 11.84DD231 pKa = 3.49ARR233 pKa = 11.84EE234 pKa = 3.3EE235 pKa = 4.1AYY237 pKa = 10.6RR238 pKa = 11.84HH239 pKa = 5.13GLEE242 pKa = 4.19SKK244 pKa = 10.25DD245 pKa = 3.62AIMAVFIKK253 pKa = 10.06TEE255 pKa = 4.07TSTSGTDD262 pKa = 3.28PRR264 pKa = 11.84NISPRR269 pKa = 11.84QLKK272 pKa = 9.59FLSVLGPFVAAIEE285 pKa = 4.27KK286 pKa = 9.59QAKK289 pKa = 8.74CCDD292 pKa = 3.66YY293 pKa = 10.87LVKK296 pKa = 10.73GKK298 pKa = 10.08NPEE301 pKa = 3.66EE302 pKa = 3.92RR303 pKa = 11.84GQYY306 pKa = 9.1ISQYY310 pKa = 7.69WRR312 pKa = 11.84HH313 pKa = 5.58SVIEE317 pKa = 4.12TDD319 pKa = 4.58FSRR322 pKa = 11.84FDD324 pKa = 3.47MTVSADD330 pKa = 3.49FVKK333 pKa = 9.86TVEE336 pKa = 3.65RR337 pKa = 11.84SMFRR341 pKa = 11.84RR342 pKa = 11.84AFPEE346 pKa = 3.94GLYY349 pKa = 10.0PALDD353 pKa = 3.68EE354 pKa = 5.3CLDD357 pKa = 4.33LLPVMTGASSLGVLYY372 pKa = 10.89SVAGTRR378 pKa = 11.84ASGDD382 pKa = 3.08AHH384 pKa = 5.82TSIANGVLNRR394 pKa = 11.84FVIWASLRR402 pKa = 11.84NMDD405 pKa = 3.78PSTWVSFHH413 pKa = 6.84EE414 pKa = 4.71GDD416 pKa = 5.21DD417 pKa = 4.75GIIHH421 pKa = 7.24CDD423 pKa = 3.22SAVTQQICDD432 pKa = 3.78YY433 pKa = 10.4LAFAAFLGFKK443 pKa = 9.99LKK445 pKa = 10.28VVRR448 pKa = 11.84PPSHH452 pKa = 5.84EE453 pKa = 3.8HH454 pKa = 6.59ANFCGRR460 pKa = 11.84HH461 pKa = 4.15TCEE464 pKa = 3.42EE465 pKa = 4.29CHH467 pKa = 7.33RR468 pKa = 11.84EE469 pKa = 4.0FCDD472 pKa = 4.78LPRR475 pKa = 11.84ALSKK479 pKa = 9.62FHH481 pKa = 6.36ITFKK485 pKa = 10.37QGEE488 pKa = 4.3PRR490 pKa = 11.84SLLLAKK496 pKa = 10.2ALSYY500 pKa = 10.4FATDD504 pKa = 2.42AHH506 pKa = 6.03TPIIGTMCDD515 pKa = 2.84ALIRR519 pKa = 11.84HH520 pKa = 6.9LSPQVSDD527 pKa = 2.79RR528 pKa = 11.84LMRR531 pKa = 11.84RR532 pKa = 11.84RR533 pKa = 11.84LHH535 pKa = 6.67AMSRR539 pKa = 11.84YY540 pKa = 10.05DD541 pKa = 3.29RR542 pKa = 11.84DD543 pKa = 3.19RR544 pKa = 11.84TIRR547 pKa = 11.84GMHH550 pKa = 5.66NTFMSPSACCRR561 pKa = 11.84AAISNVCGWDD571 pKa = 3.37PKK573 pKa = 11.14LQVAWEE579 pKa = 4.06AQLATWRR586 pKa = 11.84YY587 pKa = 8.88GITEE591 pKa = 4.34ISPLSVEE598 pKa = 4.24DD599 pKa = 4.26HH600 pKa = 6.94LVDD603 pKa = 3.64TDD605 pKa = 4.36SVTIYY610 pKa = 10.5QGG612 pKa = 2.97
Molecular weight: 69.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFE3|A0A1L3KFE3_9VIRU RNA-directed RNA polymerase OS=Wenzhou tombus-like virus 18 OX=1923671 PE=4 SV=1
MM1 pKa = 7.23TGSNRR6 pKa = 11.84RR7 pKa = 11.84ANAGRR12 pKa = 11.84KK13 pKa = 6.07TQPKK17 pKa = 8.16PQRR20 pKa = 11.84KK21 pKa = 7.74PRR23 pKa = 11.84APRR26 pKa = 11.84RR27 pKa = 11.84PKK29 pKa = 9.62VQNAPRR35 pKa = 11.84IQQGGGPVPLLEE47 pKa = 5.01SNSNMRR53 pKa = 11.84QMHH56 pKa = 6.27NGMTRR61 pKa = 11.84VVGSDD66 pKa = 3.35YY67 pKa = 11.28LGVVSVAGNPADD79 pKa = 3.55AAAKK83 pKa = 8.57VRR85 pKa = 11.84KK86 pKa = 8.96VLSVSPSSFPGTRR99 pKa = 11.84LTQMSDD105 pKa = 2.08LWEE108 pKa = 4.37RR109 pKa = 11.84YY110 pKa = 6.56VFRR113 pKa = 11.84QFRR116 pKa = 11.84VRR118 pKa = 11.84YY119 pKa = 8.61VPSVPNTLACQVMVYY134 pKa = 10.46QDD136 pKa = 4.5TDD138 pKa = 3.66PQDD141 pKa = 3.88DD142 pKa = 3.65PTAIKK147 pKa = 10.74DD148 pKa = 3.26ADD150 pKa = 3.66ALLRR154 pKa = 11.84QATAQTGSQQWNFNSAKK171 pKa = 10.1VIHH174 pKa = 6.06LAKK177 pKa = 10.57RR178 pKa = 11.84SDD180 pKa = 3.3NQLYY184 pKa = 7.83YY185 pKa = 10.29TGPVKK190 pKa = 10.5EE191 pKa = 4.26NPRR194 pKa = 11.84FNQQGVVYY202 pKa = 8.22FIQVSQALDD211 pKa = 3.49MNGKK215 pKa = 8.74PLTADD220 pKa = 3.59MEE222 pKa = 4.82CGSLYY227 pKa = 10.58VDD229 pKa = 3.55WVIDD233 pKa = 3.84FQTPQVNPSAVEE245 pKa = 3.6ARR247 pKa = 11.84LPSAGFFTRR256 pKa = 11.84QILVNDD262 pKa = 4.04KK263 pKa = 9.76LTFAAQASLTVALTALSATPKK284 pKa = 10.05VWLEE288 pKa = 3.68GPTRR292 pKa = 11.84VDD294 pKa = 3.4LATLGGPGGNLVQIEE309 pKa = 4.35LTRR312 pKa = 11.84AQPGDD317 pKa = 3.65YY318 pKa = 8.83TVKK321 pKa = 10.46FVGCDD326 pKa = 3.01SVTLVSSAPIAA337 pKa = 3.82
MM1 pKa = 7.23TGSNRR6 pKa = 11.84RR7 pKa = 11.84ANAGRR12 pKa = 11.84KK13 pKa = 6.07TQPKK17 pKa = 8.16PQRR20 pKa = 11.84KK21 pKa = 7.74PRR23 pKa = 11.84APRR26 pKa = 11.84RR27 pKa = 11.84PKK29 pKa = 9.62VQNAPRR35 pKa = 11.84IQQGGGPVPLLEE47 pKa = 5.01SNSNMRR53 pKa = 11.84QMHH56 pKa = 6.27NGMTRR61 pKa = 11.84VVGSDD66 pKa = 3.35YY67 pKa = 11.28LGVVSVAGNPADD79 pKa = 3.55AAAKK83 pKa = 8.57VRR85 pKa = 11.84KK86 pKa = 8.96VLSVSPSSFPGTRR99 pKa = 11.84LTQMSDD105 pKa = 2.08LWEE108 pKa = 4.37RR109 pKa = 11.84YY110 pKa = 6.56VFRR113 pKa = 11.84QFRR116 pKa = 11.84VRR118 pKa = 11.84YY119 pKa = 8.61VPSVPNTLACQVMVYY134 pKa = 10.46QDD136 pKa = 4.5TDD138 pKa = 3.66PQDD141 pKa = 3.88DD142 pKa = 3.65PTAIKK147 pKa = 10.74DD148 pKa = 3.26ADD150 pKa = 3.66ALLRR154 pKa = 11.84QATAQTGSQQWNFNSAKK171 pKa = 10.1VIHH174 pKa = 6.06LAKK177 pKa = 10.57RR178 pKa = 11.84SDD180 pKa = 3.3NQLYY184 pKa = 7.83YY185 pKa = 10.29TGPVKK190 pKa = 10.5EE191 pKa = 4.26NPRR194 pKa = 11.84FNQQGVVYY202 pKa = 8.22FIQVSQALDD211 pKa = 3.49MNGKK215 pKa = 8.74PLTADD220 pKa = 3.59MEE222 pKa = 4.82CGSLYY227 pKa = 10.58VDD229 pKa = 3.55WVIDD233 pKa = 3.84FQTPQVNPSAVEE245 pKa = 3.6ARR247 pKa = 11.84LPSAGFFTRR256 pKa = 11.84QILVNDD262 pKa = 4.04KK263 pKa = 9.76LTFAAQASLTVALTALSATPKK284 pKa = 10.05VWLEE288 pKa = 3.68GPTRR292 pKa = 11.84VDD294 pKa = 3.4LATLGGPGGNLVQIEE309 pKa = 4.35LTRR312 pKa = 11.84AQPGDD317 pKa = 3.65YY318 pKa = 8.83TVKK321 pKa = 10.46FVGCDD326 pKa = 3.01SVTLVSSAPIAA337 pKa = 3.82
Molecular weight: 36.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1389 |
337 |
612 |
463.0 |
51.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.423 ± 0.495 | 2.376 ± 0.442 |
5.688 ± 0.288 | 3.6 ± 0.882 |
3.384 ± 0.633 | 5.904 ± 0.635 |
2.592 ± 0.697 | 2.808 ± 0.95 |
3.816 ± 0.127 | 9.071 ± 0.513 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.016 ± 0.356 | 3.168 ± 0.59 |
7.703 ± 0.091 | 4.824 ± 0.965 |
7.703 ± 0.308 | 7.703 ± 0.308 |
7.055 ± 0.789 | 7.991 ± 0.973 |
1.872 ± 0.21 | 2.304 ± 0.186 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
