
Ferrimonas balearica (strain DSM 9799 / CCM 4581 / KCTC 23876 / PAT)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Ferrimonadaceae; Ferrimonas; Ferrimonas balearica
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
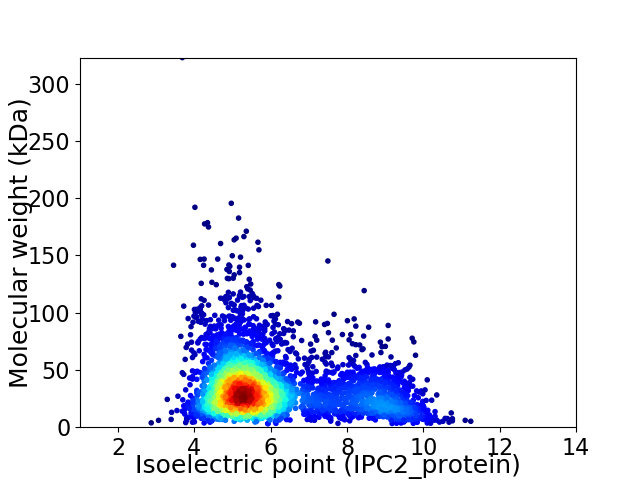
Virtual 2D-PAGE plot for 3781 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
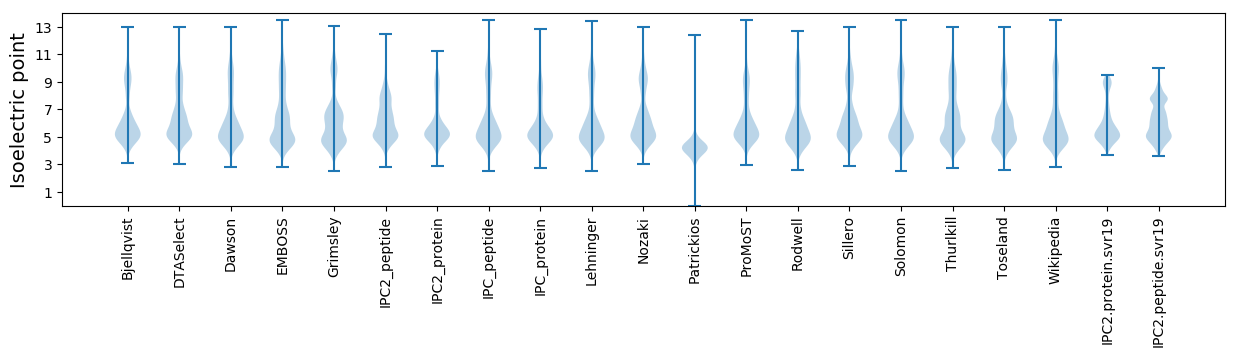
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E1SU05|E1SU05_FERBD Uncharacterized protein OS=Ferrimonas balearica (strain DSM 9799 / CCM 4581 / KCTC 23876 / PAT) OX=550540 GN=Fbal_3049 PE=4 SV=1
MM1 pKa = 8.32RR2 pKa = 11.84YY3 pKa = 9.02PLLASAVLLALTGCGSGSDD22 pKa = 3.68SDD24 pKa = 4.55SGNTPDD30 pKa = 4.85PTPQPTPNTAPTLSLSQYY48 pKa = 6.97TTAEE52 pKa = 4.08DD53 pKa = 4.28TPFSLSLADD62 pKa = 4.53LVSDD66 pKa = 3.87VDD68 pKa = 3.8GDD70 pKa = 4.2NISLSVEE77 pKa = 3.75ACGDD81 pKa = 3.49QLSCQLSGTTLTLTPADD98 pKa = 4.92DD99 pKa = 3.75YY100 pKa = 11.56HH101 pKa = 8.5GEE103 pKa = 3.93QSVRR107 pKa = 11.84LSASDD112 pKa = 4.29GEE114 pKa = 4.45ASTSAEE120 pKa = 3.75LTLTVEE126 pKa = 4.71AVNDD130 pKa = 3.86APLLAVSDD138 pKa = 3.91QQVEE142 pKa = 4.23EE143 pKa = 4.96DD144 pKa = 4.14GSLTLDD150 pKa = 3.7LASWISDD157 pKa = 3.19VDD159 pKa = 3.64GDD161 pKa = 4.58ALTLSVASCAANIDD175 pKa = 3.86CALDD179 pKa = 3.7NTSLTLTPAADD190 pKa = 3.55HH191 pKa = 6.79FGDD194 pKa = 3.62SHH196 pKa = 8.75SIGLTVTDD204 pKa = 4.35AANASTSASFNLTVSGVNDD223 pKa = 3.63APQWQAIAAQTLQYY237 pKa = 10.54DD238 pKa = 3.5QSLTLDD244 pKa = 4.24LTRR247 pKa = 11.84HH248 pKa = 5.87IADD251 pKa = 4.48ADD253 pKa = 3.76NDD255 pKa = 4.18SLSFSVVSCDD265 pKa = 4.72NLQCQLNGSEE275 pKa = 4.63LTLSAPAEE283 pKa = 4.27AGVTYY288 pKa = 10.24PLTLRR293 pKa = 11.84ATDD296 pKa = 4.05GSDD299 pKa = 3.61EE300 pKa = 4.3VDD302 pKa = 3.06TALQVAITKK311 pKa = 9.9VPVDD315 pKa = 4.97AIAGVQFFPPYY326 pKa = 10.33NGLTGVDD333 pKa = 3.61NVTLSLHH340 pKa = 6.07SEE342 pKa = 3.85QDD344 pKa = 3.45LTQLTINGEE353 pKa = 4.46VITLEE358 pKa = 5.28NPRR361 pKa = 11.84QWQQNFPLQMGDD373 pKa = 3.37TLFTVVLEE381 pKa = 4.5TATEE385 pKa = 4.17TLSLTKK391 pKa = 9.46TVRR394 pKa = 11.84YY395 pKa = 9.9QGDD398 pKa = 3.76LHH400 pKa = 7.13QGSADD405 pKa = 3.41LTYY408 pKa = 11.0LGDD411 pKa = 3.64RR412 pKa = 11.84TVLVDD417 pKa = 4.4DD418 pKa = 4.28YY419 pKa = 11.59RR420 pKa = 11.84AGALVAFDD428 pKa = 4.46LDD430 pKa = 3.92TRR432 pKa = 11.84EE433 pKa = 4.04TTAYY437 pKa = 9.84PSDD440 pKa = 3.95RR441 pKa = 11.84LTGMNIDD448 pKa = 3.6FLDD451 pKa = 3.72WYY453 pKa = 10.52VALDD457 pKa = 3.55SEE459 pKa = 4.72RR460 pKa = 11.84LLGSAWDD467 pKa = 3.63QDD469 pKa = 3.34AAANKK474 pKa = 9.62LVEE477 pKa = 4.41VNLTDD482 pKa = 3.41LTITEE487 pKa = 5.09RR488 pKa = 11.84YY489 pKa = 8.88NLSNVPVQVSHH500 pKa = 6.49LTASTDD506 pKa = 2.9ASLVYY511 pKa = 9.42FVFDD515 pKa = 5.15NSRR518 pKa = 11.84QDD520 pKa = 3.62PNTGAWYY527 pKa = 8.77PVEE530 pKa = 4.31EE531 pKa = 4.93FYY533 pKa = 11.15QFEE536 pKa = 4.53VATNTLTLLADD547 pKa = 4.12TAQVDD552 pKa = 3.84DD553 pKa = 5.56DD554 pKa = 4.68GPLTRR559 pKa = 11.84VRR561 pKa = 11.84GMILDD566 pKa = 3.83EE567 pKa = 4.5TNDD570 pKa = 3.48RR571 pKa = 11.84LLMVNQNYY579 pKa = 10.1SDD581 pKa = 3.7PAARR585 pKa = 11.84LLALPLSGPDD595 pKa = 3.02EE596 pKa = 4.42GRR598 pKa = 11.84LVEE601 pKa = 4.64LTPSSDD607 pKa = 3.46PGCLDD612 pKa = 3.98FAAQEE617 pKa = 4.26YY618 pKa = 10.16YY619 pKa = 10.53EE620 pKa = 4.38LRR622 pKa = 11.84NVRR625 pKa = 11.84DD626 pKa = 4.38GKK628 pKa = 10.13WLMVMQNDD636 pKa = 3.15NWASYY641 pKa = 10.6QLDD644 pKa = 3.83INTLCLTEE652 pKa = 3.78HH653 pKa = 6.36HH654 pKa = 6.69RR655 pKa = 11.84VPEE658 pKa = 4.02SLLLNRR664 pKa = 11.84EE665 pKa = 4.13SLQGIDD671 pKa = 3.68VNPLTGEE678 pKa = 4.06ILLGFYY684 pKa = 9.57PGPIVYY690 pKa = 9.92QPEE693 pKa = 3.87TDD695 pKa = 3.21SYY697 pKa = 11.26RR698 pKa = 11.84GAEE701 pKa = 3.82AAGFAGEE708 pKa = 4.15PWNIQEE714 pKa = 4.38PTAIHH719 pKa = 5.85YY720 pKa = 9.45RR721 pKa = 11.84ADD723 pKa = 3.18THH725 pKa = 6.55QIVYY729 pKa = 10.79GDD731 pKa = 4.37DD732 pKa = 3.39QFLMQLDD739 pKa = 4.32LQTGEE744 pKa = 4.31VTQLAQLRR752 pKa = 11.84DD753 pKa = 4.13NILKK757 pKa = 10.62LEE759 pKa = 4.04WDD761 pKa = 3.93EE762 pKa = 4.51ASQRR766 pKa = 11.84YY767 pKa = 8.28YY768 pKa = 11.07LYY770 pKa = 10.77DD771 pKa = 3.76EE772 pKa = 4.62YY773 pKa = 11.43DD774 pKa = 3.21GLLAYY779 pKa = 10.07DD780 pKa = 3.58VAGEE784 pKa = 3.92QLVTLVEE791 pKa = 4.31EE792 pKa = 4.64ASGQHH797 pKa = 5.83FSYY800 pKa = 10.27PALASDD806 pKa = 3.63GQLYY810 pKa = 10.01YY811 pKa = 10.8VDD813 pKa = 4.91SDD815 pKa = 3.64VLNRR819 pKa = 11.84INPEE823 pKa = 3.72TLEE826 pKa = 4.16INTVSPHH833 pKa = 6.84DD834 pKa = 4.11DD835 pKa = 3.13AAGFRR840 pKa = 11.84SITSLHH846 pKa = 6.55LDD848 pKa = 3.54EE849 pKa = 5.69PNNRR853 pKa = 11.84AIISVSSGGDD863 pKa = 3.08TTYY866 pKa = 10.75IGLDD870 pKa = 3.24LDD872 pKa = 3.88SGVRR876 pKa = 11.84TILLPEE882 pKa = 4.51SDD884 pKa = 5.34LNDD887 pKa = 3.22GDD889 pKa = 4.58YY890 pKa = 11.35NILSADD896 pKa = 4.02GSLMWYY902 pKa = 9.99FDD904 pKa = 3.6WANDD908 pKa = 3.5QVMSLNLDD916 pKa = 3.59TQAITPVTPASTNHH930 pKa = 5.78YY931 pKa = 10.33KK932 pKa = 10.87LGDD935 pKa = 3.49PTGLFRR941 pKa = 11.84LSDD944 pKa = 5.27DD945 pKa = 3.66ILLMGDD951 pKa = 3.78DD952 pKa = 4.49DD953 pKa = 6.52FGGFWMINLTTDD965 pKa = 2.78EE966 pKa = 4.32KK967 pKa = 11.28LLYY970 pKa = 10.19RR971 pKa = 4.69
MM1 pKa = 8.32RR2 pKa = 11.84YY3 pKa = 9.02PLLASAVLLALTGCGSGSDD22 pKa = 3.68SDD24 pKa = 4.55SGNTPDD30 pKa = 4.85PTPQPTPNTAPTLSLSQYY48 pKa = 6.97TTAEE52 pKa = 4.08DD53 pKa = 4.28TPFSLSLADD62 pKa = 4.53LVSDD66 pKa = 3.87VDD68 pKa = 3.8GDD70 pKa = 4.2NISLSVEE77 pKa = 3.75ACGDD81 pKa = 3.49QLSCQLSGTTLTLTPADD98 pKa = 4.92DD99 pKa = 3.75YY100 pKa = 11.56HH101 pKa = 8.5GEE103 pKa = 3.93QSVRR107 pKa = 11.84LSASDD112 pKa = 4.29GEE114 pKa = 4.45ASTSAEE120 pKa = 3.75LTLTVEE126 pKa = 4.71AVNDD130 pKa = 3.86APLLAVSDD138 pKa = 3.91QQVEE142 pKa = 4.23EE143 pKa = 4.96DD144 pKa = 4.14GSLTLDD150 pKa = 3.7LASWISDD157 pKa = 3.19VDD159 pKa = 3.64GDD161 pKa = 4.58ALTLSVASCAANIDD175 pKa = 3.86CALDD179 pKa = 3.7NTSLTLTPAADD190 pKa = 3.55HH191 pKa = 6.79FGDD194 pKa = 3.62SHH196 pKa = 8.75SIGLTVTDD204 pKa = 4.35AANASTSASFNLTVSGVNDD223 pKa = 3.63APQWQAIAAQTLQYY237 pKa = 10.54DD238 pKa = 3.5QSLTLDD244 pKa = 4.24LTRR247 pKa = 11.84HH248 pKa = 5.87IADD251 pKa = 4.48ADD253 pKa = 3.76NDD255 pKa = 4.18SLSFSVVSCDD265 pKa = 4.72NLQCQLNGSEE275 pKa = 4.63LTLSAPAEE283 pKa = 4.27AGVTYY288 pKa = 10.24PLTLRR293 pKa = 11.84ATDD296 pKa = 4.05GSDD299 pKa = 3.61EE300 pKa = 4.3VDD302 pKa = 3.06TALQVAITKK311 pKa = 9.9VPVDD315 pKa = 4.97AIAGVQFFPPYY326 pKa = 10.33NGLTGVDD333 pKa = 3.61NVTLSLHH340 pKa = 6.07SEE342 pKa = 3.85QDD344 pKa = 3.45LTQLTINGEE353 pKa = 4.46VITLEE358 pKa = 5.28NPRR361 pKa = 11.84QWQQNFPLQMGDD373 pKa = 3.37TLFTVVLEE381 pKa = 4.5TATEE385 pKa = 4.17TLSLTKK391 pKa = 9.46TVRR394 pKa = 11.84YY395 pKa = 9.9QGDD398 pKa = 3.76LHH400 pKa = 7.13QGSADD405 pKa = 3.41LTYY408 pKa = 11.0LGDD411 pKa = 3.64RR412 pKa = 11.84TVLVDD417 pKa = 4.4DD418 pKa = 4.28YY419 pKa = 11.59RR420 pKa = 11.84AGALVAFDD428 pKa = 4.46LDD430 pKa = 3.92TRR432 pKa = 11.84EE433 pKa = 4.04TTAYY437 pKa = 9.84PSDD440 pKa = 3.95RR441 pKa = 11.84LTGMNIDD448 pKa = 3.6FLDD451 pKa = 3.72WYY453 pKa = 10.52VALDD457 pKa = 3.55SEE459 pKa = 4.72RR460 pKa = 11.84LLGSAWDD467 pKa = 3.63QDD469 pKa = 3.34AAANKK474 pKa = 9.62LVEE477 pKa = 4.41VNLTDD482 pKa = 3.41LTITEE487 pKa = 5.09RR488 pKa = 11.84YY489 pKa = 8.88NLSNVPVQVSHH500 pKa = 6.49LTASTDD506 pKa = 2.9ASLVYY511 pKa = 9.42FVFDD515 pKa = 5.15NSRR518 pKa = 11.84QDD520 pKa = 3.62PNTGAWYY527 pKa = 8.77PVEE530 pKa = 4.31EE531 pKa = 4.93FYY533 pKa = 11.15QFEE536 pKa = 4.53VATNTLTLLADD547 pKa = 4.12TAQVDD552 pKa = 3.84DD553 pKa = 5.56DD554 pKa = 4.68GPLTRR559 pKa = 11.84VRR561 pKa = 11.84GMILDD566 pKa = 3.83EE567 pKa = 4.5TNDD570 pKa = 3.48RR571 pKa = 11.84LLMVNQNYY579 pKa = 10.1SDD581 pKa = 3.7PAARR585 pKa = 11.84LLALPLSGPDD595 pKa = 3.02EE596 pKa = 4.42GRR598 pKa = 11.84LVEE601 pKa = 4.64LTPSSDD607 pKa = 3.46PGCLDD612 pKa = 3.98FAAQEE617 pKa = 4.26YY618 pKa = 10.16YY619 pKa = 10.53EE620 pKa = 4.38LRR622 pKa = 11.84NVRR625 pKa = 11.84DD626 pKa = 4.38GKK628 pKa = 10.13WLMVMQNDD636 pKa = 3.15NWASYY641 pKa = 10.6QLDD644 pKa = 3.83INTLCLTEE652 pKa = 3.78HH653 pKa = 6.36HH654 pKa = 6.69RR655 pKa = 11.84VPEE658 pKa = 4.02SLLLNRR664 pKa = 11.84EE665 pKa = 4.13SLQGIDD671 pKa = 3.68VNPLTGEE678 pKa = 4.06ILLGFYY684 pKa = 9.57PGPIVYY690 pKa = 9.92QPEE693 pKa = 3.87TDD695 pKa = 3.21SYY697 pKa = 11.26RR698 pKa = 11.84GAEE701 pKa = 3.82AAGFAGEE708 pKa = 4.15PWNIQEE714 pKa = 4.38PTAIHH719 pKa = 5.85YY720 pKa = 9.45RR721 pKa = 11.84ADD723 pKa = 3.18THH725 pKa = 6.55QIVYY729 pKa = 10.79GDD731 pKa = 4.37DD732 pKa = 3.39QFLMQLDD739 pKa = 4.32LQTGEE744 pKa = 4.31VTQLAQLRR752 pKa = 11.84DD753 pKa = 4.13NILKK757 pKa = 10.62LEE759 pKa = 4.04WDD761 pKa = 3.93EE762 pKa = 4.51ASQRR766 pKa = 11.84YY767 pKa = 8.28YY768 pKa = 11.07LYY770 pKa = 10.77DD771 pKa = 3.76EE772 pKa = 4.62YY773 pKa = 11.43DD774 pKa = 3.21GLLAYY779 pKa = 10.07DD780 pKa = 3.58VAGEE784 pKa = 3.92QLVTLVEE791 pKa = 4.31EE792 pKa = 4.64ASGQHH797 pKa = 5.83FSYY800 pKa = 10.27PALASDD806 pKa = 3.63GQLYY810 pKa = 10.01YY811 pKa = 10.8VDD813 pKa = 4.91SDD815 pKa = 3.64VLNRR819 pKa = 11.84INPEE823 pKa = 3.72TLEE826 pKa = 4.16INTVSPHH833 pKa = 6.84DD834 pKa = 4.11DD835 pKa = 3.13AAGFRR840 pKa = 11.84SITSLHH846 pKa = 6.55LDD848 pKa = 3.54EE849 pKa = 5.69PNNRR853 pKa = 11.84AIISVSSGGDD863 pKa = 3.08TTYY866 pKa = 10.75IGLDD870 pKa = 3.24LDD872 pKa = 3.88SGVRR876 pKa = 11.84TILLPEE882 pKa = 4.51SDD884 pKa = 5.34LNDD887 pKa = 3.22GDD889 pKa = 4.58YY890 pKa = 11.35NILSADD896 pKa = 4.02GSLMWYY902 pKa = 9.99FDD904 pKa = 3.6WANDD908 pKa = 3.5QVMSLNLDD916 pKa = 3.59TQAITPVTPASTNHH930 pKa = 5.78YY931 pKa = 10.33KK932 pKa = 10.87LGDD935 pKa = 3.49PTGLFRR941 pKa = 11.84LSDD944 pKa = 5.27DD945 pKa = 3.66ILLMGDD951 pKa = 3.78DD952 pKa = 4.49DD953 pKa = 6.52FGGFWMINLTTDD965 pKa = 2.78EE966 pKa = 4.32KK967 pKa = 11.28LLYY970 pKa = 10.19RR971 pKa = 4.69
Molecular weight: 105.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1SRV5|E1SRV5_FERBD DNA mismatch repair protein MutH OS=Ferrimonas balearica (strain DSM 9799 / CCM 4581 / KCTC 23876 / PAT) OX=550540 GN=mutH PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.39GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.39GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1264585 |
30 |
3058 |
334.5 |
36.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.712 ± 0.059 | 1.051 ± 0.016 |
5.671 ± 0.037 | 6.01 ± 0.039 |
3.54 ± 0.025 | 7.826 ± 0.035 |
2.378 ± 0.021 | 4.624 ± 0.029 |
3.377 ± 0.033 | 11.46 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.596 ± 0.019 | 3.202 ± 0.027 |
4.78 ± 0.03 | 4.942 ± 0.038 |
5.934 ± 0.035 | 5.591 ± 0.031 |
5.035 ± 0.027 | 7.067 ± 0.036 |
1.446 ± 0.016 | 2.757 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
