
Rhizoctonia mitovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.56
Get precalculated fractions of proteins
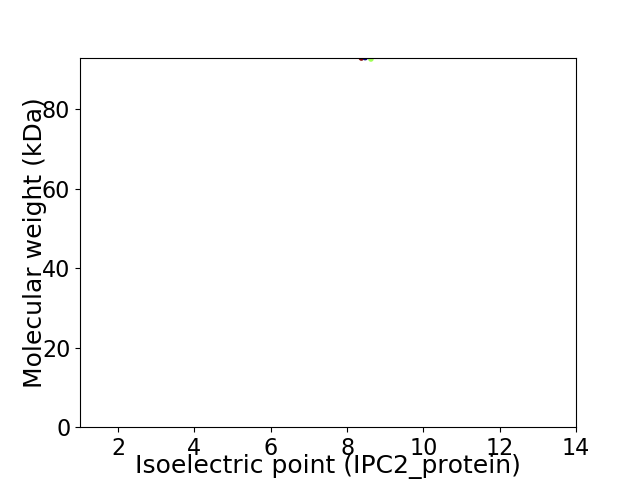
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A494RIS4|A0A494RIS4_9VIRU RNA-dependent RNA polymerase OS=Rhizoctonia mitovirus 1 OX=2421278 PE=4 SV=1
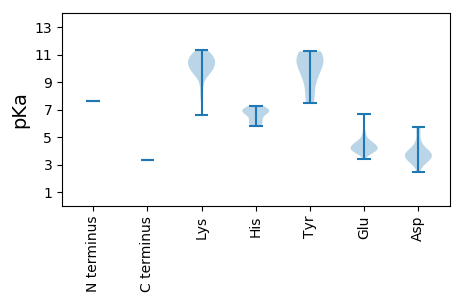
MM1 pKa = 7.59KK2 pKa = 10.68LFTTTTKK9 pKa = 10.95AVISKK14 pKa = 6.57TTTLFTRR21 pKa = 11.84SIYY24 pKa = 10.02TSHH27 pKa = 6.92TLLNTTKK34 pKa = 9.98TIEE37 pKa = 4.19SLGLRR42 pKa = 11.84GWQPMVPSVNEE53 pKa = 3.37ARR55 pKa = 11.84NNKK58 pKa = 9.61RR59 pKa = 11.84IANAISHH66 pKa = 6.69LHH68 pKa = 6.24LFMEE72 pKa = 4.82TQTGLSNPAVMALTDD87 pKa = 5.73LITEE91 pKa = 4.63LRR93 pKa = 11.84NEE95 pKa = 4.15YY96 pKa = 10.57NEE98 pKa = 4.35NPLQFVRR105 pKa = 11.84LSKK108 pKa = 11.16LMGNWFKK115 pKa = 11.34INIFQLKK122 pKa = 7.86RR123 pKa = 11.84TQGLIPVNMWDD134 pKa = 3.61SYY136 pKa = 9.32TNVPRR141 pKa = 11.84GLVGIFSAFNQLSTDD156 pKa = 3.37RR157 pKa = 11.84EE158 pKa = 3.96RR159 pKa = 11.84IIFIRR164 pKa = 11.84MVLSILDD171 pKa = 4.01LYY173 pKa = 10.75RR174 pKa = 11.84VVVTPTKK181 pKa = 9.87PDD183 pKa = 2.93LGTIVNPGTDD193 pKa = 2.44IGGIIKK199 pKa = 10.31DD200 pKa = 4.01DD201 pKa = 4.76EE202 pKa = 4.39IQKK205 pKa = 10.1SLEE208 pKa = 3.92MLGIDD213 pKa = 5.44CDD215 pKa = 4.12LLTNNFKK222 pKa = 10.46FANKK226 pKa = 9.98AVFFHH231 pKa = 6.87SSSAQGPNGQAVWNSHH247 pKa = 6.04LDD249 pKa = 3.69AKK251 pKa = 11.1ALLADD256 pKa = 3.99KK257 pKa = 10.98DD258 pKa = 3.87LFEE261 pKa = 5.23NVQQLALLLEE271 pKa = 4.7RR272 pKa = 11.84KK273 pKa = 9.88DD274 pKa = 3.64LLTVLEE280 pKa = 4.65EE281 pKa = 4.05AAKK284 pKa = 10.76LPDD287 pKa = 4.63LISTNEE293 pKa = 3.85EE294 pKa = 3.86PLKK297 pKa = 10.57HH298 pKa = 6.35SKK300 pKa = 10.26LHH302 pKa = 6.82IIFEE306 pKa = 4.7NGDD309 pKa = 3.29KK310 pKa = 10.52VRR312 pKa = 11.84PVAIIDD318 pKa = 3.87YY319 pKa = 7.47FTQEE323 pKa = 4.36LLSPFHH329 pKa = 7.06DD330 pKa = 4.01LVAGILRR337 pKa = 11.84SIPQDD342 pKa = 3.02GTFNQNAIASKK353 pKa = 10.19VKK355 pKa = 10.46EE356 pKa = 4.34FTATAGNSLFSFDD369 pKa = 3.86LTAATDD375 pKa = 3.88RR376 pKa = 11.84LPVILQRR383 pKa = 11.84RR384 pKa = 11.84IISHH388 pKa = 7.02IIKK391 pKa = 9.6IDD393 pKa = 3.31RR394 pKa = 11.84FALLWQKK401 pKa = 10.79VLTFRR406 pKa = 11.84DD407 pKa = 4.43FSLGNGHH414 pKa = 5.86SVRR417 pKa = 11.84YY418 pKa = 9.36AVGQPMGAKK427 pKa = 9.91SSFPMLGLTHH437 pKa = 6.91HH438 pKa = 7.24IIVQIAALRR447 pKa = 11.84VGFSTMFKK455 pKa = 10.46DD456 pKa = 4.32YY457 pKa = 11.23VILGDD462 pKa = 5.23DD463 pKa = 3.65IMIANEE469 pKa = 4.0KK470 pKa = 10.36VATQYY475 pKa = 10.93RR476 pKa = 11.84RR477 pKa = 11.84IMEE480 pKa = 4.31SLGLAISQHH489 pKa = 5.83KK490 pKa = 10.43SIISTNSTTSAQIAEE505 pKa = 3.84ICRR508 pKa = 11.84RR509 pKa = 11.84VFVRR513 pKa = 11.84GIEE516 pKa = 4.01ITPLPMKK523 pKa = 10.69LIANVFEE530 pKa = 4.51NGSMFLQLQEE540 pKa = 4.32KK541 pKa = 8.53LTEE544 pKa = 4.58RR545 pKa = 11.84GLVFAPEE552 pKa = 3.9NFEE555 pKa = 4.56LLVAGFKK562 pKa = 10.89LSLQDD567 pKa = 3.4FNQVVLQNSLPQWLTKK583 pKa = 9.67LTEE586 pKa = 4.17TFSFEE591 pKa = 6.65DD592 pKa = 3.27SDD594 pKa = 3.45EE595 pKa = 4.44WNFKK599 pKa = 8.13TWTDD603 pKa = 3.49VGIDD607 pKa = 3.58EE608 pKa = 5.09QMLLEE613 pKa = 4.36FWKK616 pKa = 9.01YY617 pKa = 8.02TVISEE622 pKa = 4.03QFKK625 pKa = 10.82RR626 pKa = 11.84ISTLIQAATNGFKK639 pKa = 10.61AISKK643 pKa = 10.16AISLGDD649 pKa = 3.31ILFAQDD655 pKa = 3.75PQTGDD660 pKa = 3.87RR661 pKa = 11.84IPAKK665 pKa = 9.94MFTLEE670 pKa = 5.5TLQQWNILKK679 pKa = 9.72VAHH682 pKa = 6.89PARR685 pKa = 11.84FVMLKK690 pKa = 9.23EE691 pKa = 3.99VEE693 pKa = 4.36RR694 pKa = 11.84ISGIMRR700 pKa = 11.84RR701 pKa = 11.84VASARR706 pKa = 11.84GAEE709 pKa = 4.26LFVSLMDD716 pKa = 5.71SIVDD720 pKa = 3.43QLKK723 pKa = 9.98YY724 pKa = 10.97SILEE728 pKa = 4.21VIKK731 pKa = 11.07DD732 pKa = 3.62KK733 pKa = 11.04DD734 pKa = 3.73LKK736 pKa = 10.48ASQMTRR742 pKa = 11.84SMIDD746 pKa = 2.8KK747 pKa = 9.31TLANMRR753 pKa = 11.84AANLAEE759 pKa = 4.07NKK761 pKa = 9.72QLSFTVKK768 pKa = 10.46LEE770 pKa = 3.93SLSIVWVLKK779 pKa = 10.06VSLNGSCTLSEE790 pKa = 4.14STMNVPTTTRR800 pKa = 11.84DD801 pKa = 3.22AKK803 pKa = 11.0AQLLKK808 pKa = 10.84YY809 pKa = 10.39RR810 pKa = 11.84SQNQSFKK817 pKa = 11.34SSFGSSKK824 pKa = 9.73STT826 pKa = 3.32
MM1 pKa = 7.59KK2 pKa = 10.68LFTTTTKK9 pKa = 10.95AVISKK14 pKa = 6.57TTTLFTRR21 pKa = 11.84SIYY24 pKa = 10.02TSHH27 pKa = 6.92TLLNTTKK34 pKa = 9.98TIEE37 pKa = 4.19SLGLRR42 pKa = 11.84GWQPMVPSVNEE53 pKa = 3.37ARR55 pKa = 11.84NNKK58 pKa = 9.61RR59 pKa = 11.84IANAISHH66 pKa = 6.69LHH68 pKa = 6.24LFMEE72 pKa = 4.82TQTGLSNPAVMALTDD87 pKa = 5.73LITEE91 pKa = 4.63LRR93 pKa = 11.84NEE95 pKa = 4.15YY96 pKa = 10.57NEE98 pKa = 4.35NPLQFVRR105 pKa = 11.84LSKK108 pKa = 11.16LMGNWFKK115 pKa = 11.34INIFQLKK122 pKa = 7.86RR123 pKa = 11.84TQGLIPVNMWDD134 pKa = 3.61SYY136 pKa = 9.32TNVPRR141 pKa = 11.84GLVGIFSAFNQLSTDD156 pKa = 3.37RR157 pKa = 11.84EE158 pKa = 3.96RR159 pKa = 11.84IIFIRR164 pKa = 11.84MVLSILDD171 pKa = 4.01LYY173 pKa = 10.75RR174 pKa = 11.84VVVTPTKK181 pKa = 9.87PDD183 pKa = 2.93LGTIVNPGTDD193 pKa = 2.44IGGIIKK199 pKa = 10.31DD200 pKa = 4.01DD201 pKa = 4.76EE202 pKa = 4.39IQKK205 pKa = 10.1SLEE208 pKa = 3.92MLGIDD213 pKa = 5.44CDD215 pKa = 4.12LLTNNFKK222 pKa = 10.46FANKK226 pKa = 9.98AVFFHH231 pKa = 6.87SSSAQGPNGQAVWNSHH247 pKa = 6.04LDD249 pKa = 3.69AKK251 pKa = 11.1ALLADD256 pKa = 3.99KK257 pKa = 10.98DD258 pKa = 3.87LFEE261 pKa = 5.23NVQQLALLLEE271 pKa = 4.7RR272 pKa = 11.84KK273 pKa = 9.88DD274 pKa = 3.64LLTVLEE280 pKa = 4.65EE281 pKa = 4.05AAKK284 pKa = 10.76LPDD287 pKa = 4.63LISTNEE293 pKa = 3.85EE294 pKa = 3.86PLKK297 pKa = 10.57HH298 pKa = 6.35SKK300 pKa = 10.26LHH302 pKa = 6.82IIFEE306 pKa = 4.7NGDD309 pKa = 3.29KK310 pKa = 10.52VRR312 pKa = 11.84PVAIIDD318 pKa = 3.87YY319 pKa = 7.47FTQEE323 pKa = 4.36LLSPFHH329 pKa = 7.06DD330 pKa = 4.01LVAGILRR337 pKa = 11.84SIPQDD342 pKa = 3.02GTFNQNAIASKK353 pKa = 10.19VKK355 pKa = 10.46EE356 pKa = 4.34FTATAGNSLFSFDD369 pKa = 3.86LTAATDD375 pKa = 3.88RR376 pKa = 11.84LPVILQRR383 pKa = 11.84RR384 pKa = 11.84IISHH388 pKa = 7.02IIKK391 pKa = 9.6IDD393 pKa = 3.31RR394 pKa = 11.84FALLWQKK401 pKa = 10.79VLTFRR406 pKa = 11.84DD407 pKa = 4.43FSLGNGHH414 pKa = 5.86SVRR417 pKa = 11.84YY418 pKa = 9.36AVGQPMGAKK427 pKa = 9.91SSFPMLGLTHH437 pKa = 6.91HH438 pKa = 7.24IIVQIAALRR447 pKa = 11.84VGFSTMFKK455 pKa = 10.46DD456 pKa = 4.32YY457 pKa = 11.23VILGDD462 pKa = 5.23DD463 pKa = 3.65IMIANEE469 pKa = 4.0KK470 pKa = 10.36VATQYY475 pKa = 10.93RR476 pKa = 11.84RR477 pKa = 11.84IMEE480 pKa = 4.31SLGLAISQHH489 pKa = 5.83KK490 pKa = 10.43SIISTNSTTSAQIAEE505 pKa = 3.84ICRR508 pKa = 11.84RR509 pKa = 11.84VFVRR513 pKa = 11.84GIEE516 pKa = 4.01ITPLPMKK523 pKa = 10.69LIANVFEE530 pKa = 4.51NGSMFLQLQEE540 pKa = 4.32KK541 pKa = 8.53LTEE544 pKa = 4.58RR545 pKa = 11.84GLVFAPEE552 pKa = 3.9NFEE555 pKa = 4.56LLVAGFKK562 pKa = 10.89LSLQDD567 pKa = 3.4FNQVVLQNSLPQWLTKK583 pKa = 9.67LTEE586 pKa = 4.17TFSFEE591 pKa = 6.65DD592 pKa = 3.27SDD594 pKa = 3.45EE595 pKa = 4.44WNFKK599 pKa = 8.13TWTDD603 pKa = 3.49VGIDD607 pKa = 3.58EE608 pKa = 5.09QMLLEE613 pKa = 4.36FWKK616 pKa = 9.01YY617 pKa = 8.02TVISEE622 pKa = 4.03QFKK625 pKa = 10.82RR626 pKa = 11.84ISTLIQAATNGFKK639 pKa = 10.61AISKK643 pKa = 10.16AISLGDD649 pKa = 3.31ILFAQDD655 pKa = 3.75PQTGDD660 pKa = 3.87RR661 pKa = 11.84IPAKK665 pKa = 9.94MFTLEE670 pKa = 5.5TLQQWNILKK679 pKa = 9.72VAHH682 pKa = 6.89PARR685 pKa = 11.84FVMLKK690 pKa = 9.23EE691 pKa = 3.99VEE693 pKa = 4.36RR694 pKa = 11.84ISGIMRR700 pKa = 11.84RR701 pKa = 11.84VASARR706 pKa = 11.84GAEE709 pKa = 4.26LFVSLMDD716 pKa = 5.71SIVDD720 pKa = 3.43QLKK723 pKa = 9.98YY724 pKa = 10.97SILEE728 pKa = 4.21VIKK731 pKa = 11.07DD732 pKa = 3.62KK733 pKa = 11.04DD734 pKa = 3.73LKK736 pKa = 10.48ASQMTRR742 pKa = 11.84SMIDD746 pKa = 2.8KK747 pKa = 9.31TLANMRR753 pKa = 11.84AANLAEE759 pKa = 4.07NKK761 pKa = 9.72QLSFTVKK768 pKa = 10.46LEE770 pKa = 3.93SLSIVWVLKK779 pKa = 10.06VSLNGSCTLSEE790 pKa = 4.14STMNVPTTTRR800 pKa = 11.84DD801 pKa = 3.22AKK803 pKa = 11.0AQLLKK808 pKa = 10.84YY809 pKa = 10.39RR810 pKa = 11.84SQNQSFKK817 pKa = 11.34SSFGSSKK824 pKa = 9.73STT826 pKa = 3.32
Molecular weight: 92.93 kDa
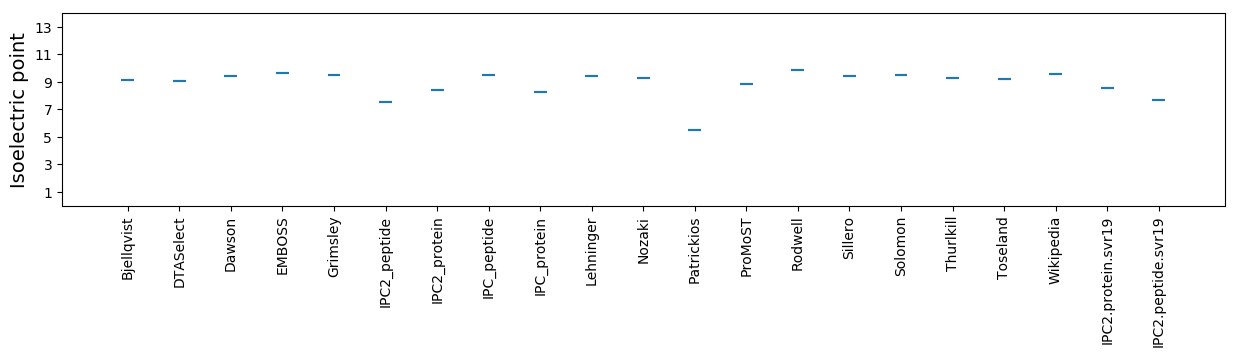
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A494RIS4|A0A494RIS4_9VIRU RNA-dependent RNA polymerase OS=Rhizoctonia mitovirus 1 OX=2421278 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.68LFTTTTKK9 pKa = 10.95AVISKK14 pKa = 6.57TTTLFTRR21 pKa = 11.84SIYY24 pKa = 10.02TSHH27 pKa = 6.92TLLNTTKK34 pKa = 9.98TIEE37 pKa = 4.19SLGLRR42 pKa = 11.84GWQPMVPSVNEE53 pKa = 3.37ARR55 pKa = 11.84NNKK58 pKa = 9.61RR59 pKa = 11.84IANAISHH66 pKa = 6.69LHH68 pKa = 6.24LFMEE72 pKa = 4.82TQTGLSNPAVMALTDD87 pKa = 5.73LITEE91 pKa = 4.63LRR93 pKa = 11.84NEE95 pKa = 4.15YY96 pKa = 10.57NEE98 pKa = 4.35NPLQFVRR105 pKa = 11.84LSKK108 pKa = 11.16LMGNWFKK115 pKa = 11.34INIFQLKK122 pKa = 7.86RR123 pKa = 11.84TQGLIPVNMWDD134 pKa = 3.61SYY136 pKa = 9.32TNVPRR141 pKa = 11.84GLVGIFSAFNQLSTDD156 pKa = 3.37RR157 pKa = 11.84EE158 pKa = 3.96RR159 pKa = 11.84IIFIRR164 pKa = 11.84MVLSILDD171 pKa = 4.01LYY173 pKa = 10.75RR174 pKa = 11.84VVVTPTKK181 pKa = 9.87PDD183 pKa = 2.93LGTIVNPGTDD193 pKa = 2.44IGGIIKK199 pKa = 10.31DD200 pKa = 4.01DD201 pKa = 4.76EE202 pKa = 4.39IQKK205 pKa = 10.1SLEE208 pKa = 3.92MLGIDD213 pKa = 5.44CDD215 pKa = 4.12LLTNNFKK222 pKa = 10.46FANKK226 pKa = 9.98AVFFHH231 pKa = 6.87SSSAQGPNGQAVWNSHH247 pKa = 6.04LDD249 pKa = 3.69AKK251 pKa = 11.1ALLADD256 pKa = 3.99KK257 pKa = 10.98DD258 pKa = 3.87LFEE261 pKa = 5.23NVQQLALLLEE271 pKa = 4.7RR272 pKa = 11.84KK273 pKa = 9.88DD274 pKa = 3.64LLTVLEE280 pKa = 4.65EE281 pKa = 4.05AAKK284 pKa = 10.76LPDD287 pKa = 4.63LISTNEE293 pKa = 3.85EE294 pKa = 3.86PLKK297 pKa = 10.57HH298 pKa = 6.35SKK300 pKa = 10.26LHH302 pKa = 6.82IIFEE306 pKa = 4.7NGDD309 pKa = 3.29KK310 pKa = 10.52VRR312 pKa = 11.84PVAIIDD318 pKa = 3.87YY319 pKa = 7.47FTQEE323 pKa = 4.36LLSPFHH329 pKa = 7.06DD330 pKa = 4.01LVAGILRR337 pKa = 11.84SIPQDD342 pKa = 3.02GTFNQNAIASKK353 pKa = 10.19VKK355 pKa = 10.46EE356 pKa = 4.34FTATAGNSLFSFDD369 pKa = 3.86LTAATDD375 pKa = 3.88RR376 pKa = 11.84LPVILQRR383 pKa = 11.84RR384 pKa = 11.84IISHH388 pKa = 7.02IIKK391 pKa = 9.6IDD393 pKa = 3.31RR394 pKa = 11.84FALLWQKK401 pKa = 10.79VLTFRR406 pKa = 11.84DD407 pKa = 4.43FSLGNGHH414 pKa = 5.86SVRR417 pKa = 11.84YY418 pKa = 9.36AVGQPMGAKK427 pKa = 9.91SSFPMLGLTHH437 pKa = 6.91HH438 pKa = 7.24IIVQIAALRR447 pKa = 11.84VGFSTMFKK455 pKa = 10.46DD456 pKa = 4.32YY457 pKa = 11.23VILGDD462 pKa = 5.23DD463 pKa = 3.65IMIANEE469 pKa = 4.0KK470 pKa = 10.36VATQYY475 pKa = 10.93RR476 pKa = 11.84RR477 pKa = 11.84IMEE480 pKa = 4.31SLGLAISQHH489 pKa = 5.83KK490 pKa = 10.43SIISTNSTTSAQIAEE505 pKa = 3.84ICRR508 pKa = 11.84RR509 pKa = 11.84VFVRR513 pKa = 11.84GIEE516 pKa = 4.01ITPLPMKK523 pKa = 10.69LIANVFEE530 pKa = 4.51NGSMFLQLQEE540 pKa = 4.32KK541 pKa = 8.53LTEE544 pKa = 4.58RR545 pKa = 11.84GLVFAPEE552 pKa = 3.9NFEE555 pKa = 4.56LLVAGFKK562 pKa = 10.89LSLQDD567 pKa = 3.4FNQVVLQNSLPQWLTKK583 pKa = 9.67LTEE586 pKa = 4.17TFSFEE591 pKa = 6.65DD592 pKa = 3.27SDD594 pKa = 3.45EE595 pKa = 4.44WNFKK599 pKa = 8.13TWTDD603 pKa = 3.49VGIDD607 pKa = 3.58EE608 pKa = 5.09QMLLEE613 pKa = 4.36FWKK616 pKa = 9.01YY617 pKa = 8.02TVISEE622 pKa = 4.03QFKK625 pKa = 10.82RR626 pKa = 11.84ISTLIQAATNGFKK639 pKa = 10.61AISKK643 pKa = 10.16AISLGDD649 pKa = 3.31ILFAQDD655 pKa = 3.75PQTGDD660 pKa = 3.87RR661 pKa = 11.84IPAKK665 pKa = 9.94MFTLEE670 pKa = 5.5TLQQWNILKK679 pKa = 9.72VAHH682 pKa = 6.89PARR685 pKa = 11.84FVMLKK690 pKa = 9.23EE691 pKa = 3.99VEE693 pKa = 4.36RR694 pKa = 11.84ISGIMRR700 pKa = 11.84RR701 pKa = 11.84VASARR706 pKa = 11.84GAEE709 pKa = 4.26LFVSLMDD716 pKa = 5.71SIVDD720 pKa = 3.43QLKK723 pKa = 9.98YY724 pKa = 10.97SILEE728 pKa = 4.21VIKK731 pKa = 11.07DD732 pKa = 3.62KK733 pKa = 11.04DD734 pKa = 3.73LKK736 pKa = 10.48ASQMTRR742 pKa = 11.84SMIDD746 pKa = 2.8KK747 pKa = 9.31TLANMRR753 pKa = 11.84AANLAEE759 pKa = 4.07NKK761 pKa = 9.72QLSFTVKK768 pKa = 10.46LEE770 pKa = 3.93SLSIVWVLKK779 pKa = 10.06VSLNGSCTLSEE790 pKa = 4.14STMNVPTTTRR800 pKa = 11.84DD801 pKa = 3.22AKK803 pKa = 11.0AQLLKK808 pKa = 10.84YY809 pKa = 10.39RR810 pKa = 11.84SQNQSFKK817 pKa = 11.34SSFGSSKK824 pKa = 9.73STT826 pKa = 3.32
MM1 pKa = 7.59KK2 pKa = 10.68LFTTTTKK9 pKa = 10.95AVISKK14 pKa = 6.57TTTLFTRR21 pKa = 11.84SIYY24 pKa = 10.02TSHH27 pKa = 6.92TLLNTTKK34 pKa = 9.98TIEE37 pKa = 4.19SLGLRR42 pKa = 11.84GWQPMVPSVNEE53 pKa = 3.37ARR55 pKa = 11.84NNKK58 pKa = 9.61RR59 pKa = 11.84IANAISHH66 pKa = 6.69LHH68 pKa = 6.24LFMEE72 pKa = 4.82TQTGLSNPAVMALTDD87 pKa = 5.73LITEE91 pKa = 4.63LRR93 pKa = 11.84NEE95 pKa = 4.15YY96 pKa = 10.57NEE98 pKa = 4.35NPLQFVRR105 pKa = 11.84LSKK108 pKa = 11.16LMGNWFKK115 pKa = 11.34INIFQLKK122 pKa = 7.86RR123 pKa = 11.84TQGLIPVNMWDD134 pKa = 3.61SYY136 pKa = 9.32TNVPRR141 pKa = 11.84GLVGIFSAFNQLSTDD156 pKa = 3.37RR157 pKa = 11.84EE158 pKa = 3.96RR159 pKa = 11.84IIFIRR164 pKa = 11.84MVLSILDD171 pKa = 4.01LYY173 pKa = 10.75RR174 pKa = 11.84VVVTPTKK181 pKa = 9.87PDD183 pKa = 2.93LGTIVNPGTDD193 pKa = 2.44IGGIIKK199 pKa = 10.31DD200 pKa = 4.01DD201 pKa = 4.76EE202 pKa = 4.39IQKK205 pKa = 10.1SLEE208 pKa = 3.92MLGIDD213 pKa = 5.44CDD215 pKa = 4.12LLTNNFKK222 pKa = 10.46FANKK226 pKa = 9.98AVFFHH231 pKa = 6.87SSSAQGPNGQAVWNSHH247 pKa = 6.04LDD249 pKa = 3.69AKK251 pKa = 11.1ALLADD256 pKa = 3.99KK257 pKa = 10.98DD258 pKa = 3.87LFEE261 pKa = 5.23NVQQLALLLEE271 pKa = 4.7RR272 pKa = 11.84KK273 pKa = 9.88DD274 pKa = 3.64LLTVLEE280 pKa = 4.65EE281 pKa = 4.05AAKK284 pKa = 10.76LPDD287 pKa = 4.63LISTNEE293 pKa = 3.85EE294 pKa = 3.86PLKK297 pKa = 10.57HH298 pKa = 6.35SKK300 pKa = 10.26LHH302 pKa = 6.82IIFEE306 pKa = 4.7NGDD309 pKa = 3.29KK310 pKa = 10.52VRR312 pKa = 11.84PVAIIDD318 pKa = 3.87YY319 pKa = 7.47FTQEE323 pKa = 4.36LLSPFHH329 pKa = 7.06DD330 pKa = 4.01LVAGILRR337 pKa = 11.84SIPQDD342 pKa = 3.02GTFNQNAIASKK353 pKa = 10.19VKK355 pKa = 10.46EE356 pKa = 4.34FTATAGNSLFSFDD369 pKa = 3.86LTAATDD375 pKa = 3.88RR376 pKa = 11.84LPVILQRR383 pKa = 11.84RR384 pKa = 11.84IISHH388 pKa = 7.02IIKK391 pKa = 9.6IDD393 pKa = 3.31RR394 pKa = 11.84FALLWQKK401 pKa = 10.79VLTFRR406 pKa = 11.84DD407 pKa = 4.43FSLGNGHH414 pKa = 5.86SVRR417 pKa = 11.84YY418 pKa = 9.36AVGQPMGAKK427 pKa = 9.91SSFPMLGLTHH437 pKa = 6.91HH438 pKa = 7.24IIVQIAALRR447 pKa = 11.84VGFSTMFKK455 pKa = 10.46DD456 pKa = 4.32YY457 pKa = 11.23VILGDD462 pKa = 5.23DD463 pKa = 3.65IMIANEE469 pKa = 4.0KK470 pKa = 10.36VATQYY475 pKa = 10.93RR476 pKa = 11.84RR477 pKa = 11.84IMEE480 pKa = 4.31SLGLAISQHH489 pKa = 5.83KK490 pKa = 10.43SIISTNSTTSAQIAEE505 pKa = 3.84ICRR508 pKa = 11.84RR509 pKa = 11.84VFVRR513 pKa = 11.84GIEE516 pKa = 4.01ITPLPMKK523 pKa = 10.69LIANVFEE530 pKa = 4.51NGSMFLQLQEE540 pKa = 4.32KK541 pKa = 8.53LTEE544 pKa = 4.58RR545 pKa = 11.84GLVFAPEE552 pKa = 3.9NFEE555 pKa = 4.56LLVAGFKK562 pKa = 10.89LSLQDD567 pKa = 3.4FNQVVLQNSLPQWLTKK583 pKa = 9.67LTEE586 pKa = 4.17TFSFEE591 pKa = 6.65DD592 pKa = 3.27SDD594 pKa = 3.45EE595 pKa = 4.44WNFKK599 pKa = 8.13TWTDD603 pKa = 3.49VGIDD607 pKa = 3.58EE608 pKa = 5.09QMLLEE613 pKa = 4.36FWKK616 pKa = 9.01YY617 pKa = 8.02TVISEE622 pKa = 4.03QFKK625 pKa = 10.82RR626 pKa = 11.84ISTLIQAATNGFKK639 pKa = 10.61AISKK643 pKa = 10.16AISLGDD649 pKa = 3.31ILFAQDD655 pKa = 3.75PQTGDD660 pKa = 3.87RR661 pKa = 11.84IPAKK665 pKa = 9.94MFTLEE670 pKa = 5.5TLQQWNILKK679 pKa = 9.72VAHH682 pKa = 6.89PARR685 pKa = 11.84FVMLKK690 pKa = 9.23EE691 pKa = 3.99VEE693 pKa = 4.36RR694 pKa = 11.84ISGIMRR700 pKa = 11.84RR701 pKa = 11.84VASARR706 pKa = 11.84GAEE709 pKa = 4.26LFVSLMDD716 pKa = 5.71SIVDD720 pKa = 3.43QLKK723 pKa = 9.98YY724 pKa = 10.97SILEE728 pKa = 4.21VIKK731 pKa = 11.07DD732 pKa = 3.62KK733 pKa = 11.04DD734 pKa = 3.73LKK736 pKa = 10.48ASQMTRR742 pKa = 11.84SMIDD746 pKa = 2.8KK747 pKa = 9.31TLANMRR753 pKa = 11.84AANLAEE759 pKa = 4.07NKK761 pKa = 9.72QLSFTVKK768 pKa = 10.46LEE770 pKa = 3.93SLSIVWVLKK779 pKa = 10.06VSLNGSCTLSEE790 pKa = 4.14STMNVPTTTRR800 pKa = 11.84DD801 pKa = 3.22AKK803 pKa = 11.0AQLLKK808 pKa = 10.84YY809 pKa = 10.39RR810 pKa = 11.84SQNQSFKK817 pKa = 11.34SSFGSSKK824 pKa = 9.73STT826 pKa = 3.32
Molecular weight: 92.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
826 |
826 |
826 |
826.0 |
92.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.78 ± 0.0 | 0.363 ± 0.0 |
4.843 ± 0.0 | 4.964 ± 0.0 |
5.69 ± 0.0 | 4.6 ± 0.0 |
1.695 ± 0.0 | 7.869 ± 0.0 |
6.295 ± 0.0 | 11.864 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.906 ± 0.0 | 5.206 ± 0.0 |
3.148 ± 0.0 | 4.843 ± 0.0 |
4.722 ± 0.0 | 7.99 ± 0.0 |
7.506 ± 0.0 | 6.053 ± 0.0 |
1.332 ± 0.0 | 1.332 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
