
Roseivivax lentus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Roseivivax
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
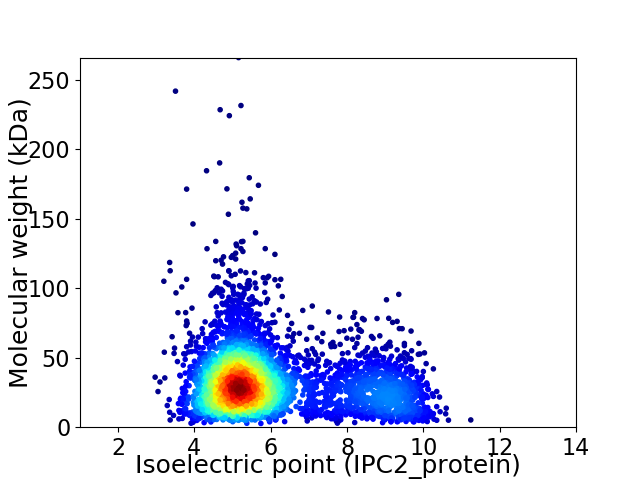
Virtual 2D-PAGE plot for 4153 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1N7P730|A0A1N7P730_9RHOB Initiator Replication protein OS=Roseivivax lentus OX=633194 GN=SAMN05421759_11341 PE=4 SV=1
MM1 pKa = 7.59ISVTTRR7 pKa = 11.84AQLEE11 pKa = 4.14NALASAQAGDD21 pKa = 4.64EE22 pKa = 4.21IVLAAGDD29 pKa = 3.82YY30 pKa = 11.36GSFDD34 pKa = 4.04FNGYY38 pKa = 9.77NYY40 pKa = 10.44SGYY43 pKa = 8.76VTVRR47 pKa = 11.84SADD50 pKa = 3.49PDD52 pKa = 3.57NPAVFDD58 pKa = 3.88QIDD61 pKa = 3.49IVGSSFLAIDD71 pKa = 3.92SVHH74 pKa = 6.58VDD76 pKa = 3.29NPSNGAASSKK86 pKa = 10.15VVNIDD91 pKa = 3.56GNSHH95 pKa = 7.43DD96 pKa = 3.85IVFSNSEE103 pKa = 4.09VNGSVTTSTNYY114 pKa = 10.49NEE116 pKa = 4.36FQGHH120 pKa = 4.8YY121 pKa = 10.51GIYY124 pKa = 9.64TGGNVRR130 pKa = 11.84NIRR133 pKa = 11.84IEE135 pKa = 4.01EE136 pKa = 4.2NEE138 pKa = 3.92VHH140 pKa = 6.74DD141 pKa = 4.31VKK143 pKa = 11.34NGFVALGSDD152 pKa = 3.76NIEE155 pKa = 3.89VVGNHH160 pKa = 6.51FDD162 pKa = 3.83RR163 pKa = 11.84LGNDD167 pKa = 3.09TMKK170 pKa = 10.56FAGVNGVLIEE180 pKa = 4.16NNIGPTSNFPSPTAHH195 pKa = 6.39VDD197 pKa = 4.61FIQFQGSSEE206 pKa = 4.3NITIRR211 pKa = 11.84GNVSLAGNIGTTQAIFMADD230 pKa = 2.88GSYY233 pKa = 11.29NNVLIEE239 pKa = 4.13QNILYY244 pKa = 9.45TGMLHH249 pKa = 7.96GITLYY254 pKa = 10.83SGANVTIRR262 pKa = 11.84DD263 pKa = 3.94NTVLNAPDD271 pKa = 4.37LVHH274 pKa = 6.97KK275 pKa = 9.77ATVIQSPDD283 pKa = 3.09GATVTGNIISATGYY297 pKa = 9.99DD298 pKa = 3.75GKK300 pKa = 11.42VSGGNLYY307 pKa = 9.69VQHH310 pKa = 6.82DD311 pKa = 4.25QPNQAHH317 pKa = 6.86HH318 pKa = 6.35YY319 pKa = 10.64SDD321 pKa = 4.15YY322 pKa = 10.88FPNAEE327 pKa = 4.11TGLGVTLQDD336 pKa = 4.12LVPLAGSLAEE346 pKa = 3.99QYY348 pKa = 10.58GAYY351 pKa = 8.96EE352 pKa = 4.04RR353 pKa = 11.84LMEE356 pKa = 5.02LLNGDD361 pKa = 3.5TWVPEE366 pKa = 4.29PAPQPDD372 pKa = 5.01PDD374 pKa = 4.52PQPEE378 pKa = 4.68PDD380 pKa = 4.23PQPLPDD386 pKa = 5.06LDD388 pKa = 4.05PAEE391 pKa = 4.33GTVFSLPGEE400 pKa = 4.12LTIADD405 pKa = 3.9AGDD408 pKa = 3.95VFEE411 pKa = 4.87MAHH414 pKa = 5.55SAEE417 pKa = 4.62LEE419 pKa = 4.09ISSGTVAFSFTADD432 pKa = 3.53DD433 pKa = 3.78VAGVQGLVAKK443 pKa = 10.14DD444 pKa = 3.1ASGYY448 pKa = 10.24VGGGHH453 pKa = 6.98HH454 pKa = 6.67FAAYY458 pKa = 10.61LEE460 pKa = 4.99GNTLMVRR467 pKa = 11.84LQDD470 pKa = 3.93AASDD474 pKa = 4.2AYY476 pKa = 11.13LSLDD480 pKa = 3.95GIEE483 pKa = 5.43AGTTYY488 pKa = 11.07DD489 pKa = 3.29VAVTFGPDD497 pKa = 3.84GGEE500 pKa = 3.45LWVNGEE506 pKa = 4.19QVSTTEE512 pKa = 5.77LVMDD516 pKa = 4.07WTQNQQHH523 pKa = 5.91LQWGGLGWGSADD535 pKa = 3.39QAAGYY540 pKa = 8.54DD541 pKa = 3.47ANFRR545 pKa = 11.84GTISDD550 pKa = 3.32ATIYY554 pKa = 8.08DD555 pKa = 3.86TKK557 pKa = 10.45LTTGGVTTQTGTEE570 pKa = 4.01FADD573 pKa = 4.09LLVGDD578 pKa = 5.24DD579 pKa = 4.02FANRR583 pKa = 11.84LQGLNGNDD591 pKa = 3.4TLNGNGGDD599 pKa = 4.05DD600 pKa = 4.18FLDD603 pKa = 4.32GGAGTDD609 pKa = 3.41RR610 pKa = 11.84VSGGAGNDD618 pKa = 3.81DD619 pKa = 4.36LDD621 pKa = 3.85VGAGNGSFQYY631 pKa = 11.39ADD633 pKa = 3.25GGAGNDD639 pKa = 3.49TYY641 pKa = 11.14HH642 pKa = 6.09YY643 pKa = 10.63AKK645 pKa = 10.67DD646 pKa = 3.26AGRR649 pKa = 11.84LYY651 pKa = 11.08LSGEE655 pKa = 4.32TASSGSADD663 pKa = 3.76RR664 pKa = 11.84IVFSDD669 pKa = 3.86LTLDD673 pKa = 4.72DD674 pKa = 4.29FTFDD678 pKa = 3.43TLDD681 pKa = 3.94YY682 pKa = 11.33GPLPYY687 pKa = 10.86GGDD690 pKa = 3.43QVALRR695 pKa = 11.84MLWNDD700 pKa = 3.67GEE702 pKa = 4.61SSGEE706 pKa = 3.72LRR708 pKa = 11.84VAA710 pKa = 4.02
MM1 pKa = 7.59ISVTTRR7 pKa = 11.84AQLEE11 pKa = 4.14NALASAQAGDD21 pKa = 4.64EE22 pKa = 4.21IVLAAGDD29 pKa = 3.82YY30 pKa = 11.36GSFDD34 pKa = 4.04FNGYY38 pKa = 9.77NYY40 pKa = 10.44SGYY43 pKa = 8.76VTVRR47 pKa = 11.84SADD50 pKa = 3.49PDD52 pKa = 3.57NPAVFDD58 pKa = 3.88QIDD61 pKa = 3.49IVGSSFLAIDD71 pKa = 3.92SVHH74 pKa = 6.58VDD76 pKa = 3.29NPSNGAASSKK86 pKa = 10.15VVNIDD91 pKa = 3.56GNSHH95 pKa = 7.43DD96 pKa = 3.85IVFSNSEE103 pKa = 4.09VNGSVTTSTNYY114 pKa = 10.49NEE116 pKa = 4.36FQGHH120 pKa = 4.8YY121 pKa = 10.51GIYY124 pKa = 9.64TGGNVRR130 pKa = 11.84NIRR133 pKa = 11.84IEE135 pKa = 4.01EE136 pKa = 4.2NEE138 pKa = 3.92VHH140 pKa = 6.74DD141 pKa = 4.31VKK143 pKa = 11.34NGFVALGSDD152 pKa = 3.76NIEE155 pKa = 3.89VVGNHH160 pKa = 6.51FDD162 pKa = 3.83RR163 pKa = 11.84LGNDD167 pKa = 3.09TMKK170 pKa = 10.56FAGVNGVLIEE180 pKa = 4.16NNIGPTSNFPSPTAHH195 pKa = 6.39VDD197 pKa = 4.61FIQFQGSSEE206 pKa = 4.3NITIRR211 pKa = 11.84GNVSLAGNIGTTQAIFMADD230 pKa = 2.88GSYY233 pKa = 11.29NNVLIEE239 pKa = 4.13QNILYY244 pKa = 9.45TGMLHH249 pKa = 7.96GITLYY254 pKa = 10.83SGANVTIRR262 pKa = 11.84DD263 pKa = 3.94NTVLNAPDD271 pKa = 4.37LVHH274 pKa = 6.97KK275 pKa = 9.77ATVIQSPDD283 pKa = 3.09GATVTGNIISATGYY297 pKa = 9.99DD298 pKa = 3.75GKK300 pKa = 11.42VSGGNLYY307 pKa = 9.69VQHH310 pKa = 6.82DD311 pKa = 4.25QPNQAHH317 pKa = 6.86HH318 pKa = 6.35YY319 pKa = 10.64SDD321 pKa = 4.15YY322 pKa = 10.88FPNAEE327 pKa = 4.11TGLGVTLQDD336 pKa = 4.12LVPLAGSLAEE346 pKa = 3.99QYY348 pKa = 10.58GAYY351 pKa = 8.96EE352 pKa = 4.04RR353 pKa = 11.84LMEE356 pKa = 5.02LLNGDD361 pKa = 3.5TWVPEE366 pKa = 4.29PAPQPDD372 pKa = 5.01PDD374 pKa = 4.52PQPEE378 pKa = 4.68PDD380 pKa = 4.23PQPLPDD386 pKa = 5.06LDD388 pKa = 4.05PAEE391 pKa = 4.33GTVFSLPGEE400 pKa = 4.12LTIADD405 pKa = 3.9AGDD408 pKa = 3.95VFEE411 pKa = 4.87MAHH414 pKa = 5.55SAEE417 pKa = 4.62LEE419 pKa = 4.09ISSGTVAFSFTADD432 pKa = 3.53DD433 pKa = 3.78VAGVQGLVAKK443 pKa = 10.14DD444 pKa = 3.1ASGYY448 pKa = 10.24VGGGHH453 pKa = 6.98HH454 pKa = 6.67FAAYY458 pKa = 10.61LEE460 pKa = 4.99GNTLMVRR467 pKa = 11.84LQDD470 pKa = 3.93AASDD474 pKa = 4.2AYY476 pKa = 11.13LSLDD480 pKa = 3.95GIEE483 pKa = 5.43AGTTYY488 pKa = 11.07DD489 pKa = 3.29VAVTFGPDD497 pKa = 3.84GGEE500 pKa = 3.45LWVNGEE506 pKa = 4.19QVSTTEE512 pKa = 5.77LVMDD516 pKa = 4.07WTQNQQHH523 pKa = 5.91LQWGGLGWGSADD535 pKa = 3.39QAAGYY540 pKa = 8.54DD541 pKa = 3.47ANFRR545 pKa = 11.84GTISDD550 pKa = 3.32ATIYY554 pKa = 8.08DD555 pKa = 3.86TKK557 pKa = 10.45LTTGGVTTQTGTEE570 pKa = 4.01FADD573 pKa = 4.09LLVGDD578 pKa = 5.24DD579 pKa = 4.02FANRR583 pKa = 11.84LQGLNGNDD591 pKa = 3.4TLNGNGGDD599 pKa = 4.05DD600 pKa = 4.18FLDD603 pKa = 4.32GGAGTDD609 pKa = 3.41RR610 pKa = 11.84VSGGAGNDD618 pKa = 3.81DD619 pKa = 4.36LDD621 pKa = 3.85VGAGNGSFQYY631 pKa = 11.39ADD633 pKa = 3.25GGAGNDD639 pKa = 3.49TYY641 pKa = 11.14HH642 pKa = 6.09YY643 pKa = 10.63AKK645 pKa = 10.67DD646 pKa = 3.26AGRR649 pKa = 11.84LYY651 pKa = 11.08LSGEE655 pKa = 4.32TASSGSADD663 pKa = 3.76RR664 pKa = 11.84IVFSDD669 pKa = 3.86LTLDD673 pKa = 4.72DD674 pKa = 4.29FTFDD678 pKa = 3.43TLDD681 pKa = 3.94YY682 pKa = 11.33GPLPYY687 pKa = 10.86GGDD690 pKa = 3.43QVALRR695 pKa = 11.84MLWNDD700 pKa = 3.67GEE702 pKa = 4.61SSGEE706 pKa = 3.72LRR708 pKa = 11.84VAA710 pKa = 4.02
Molecular weight: 74.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N7M9Q7|A0A1N7M9Q7_9RHOB L-glutamine ABC transporter membrane protein /L-glutamate ABC transporter membrane protein /L-aspartate ABC transporter membrane protein /L-asparagine ABC transporter membrane protein OS=Roseivivax lentus OX=633194 GN=SAMN05421759_104115 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84KK14 pKa = 9.18RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.42GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37AGRR29 pKa = 11.84KK30 pKa = 8.54ILNARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.07SLSAA45 pKa = 3.93
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84KK14 pKa = 9.18RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.42GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37AGRR29 pKa = 11.84KK30 pKa = 8.54ILNARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.07SLSAA45 pKa = 3.93
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1304865 |
24 |
2538 |
314.2 |
34.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.141 ± 0.063 | 0.869 ± 0.013 |
6.212 ± 0.034 | 6.045 ± 0.038 |
3.71 ± 0.025 | 8.802 ± 0.042 |
2.017 ± 0.017 | 5.005 ± 0.03 |
2.755 ± 0.027 | 10.046 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.675 ± 0.022 | 2.34 ± 0.02 |
5.31 ± 0.028 | 2.93 ± 0.019 |
7.126 ± 0.043 | 4.934 ± 0.026 |
5.433 ± 0.029 | 7.079 ± 0.029 |
1.417 ± 0.017 | 2.155 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
