
Magnetospirillum aberrantis SpK
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Magnetospirillum; Magnetospirillum aberrantis
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
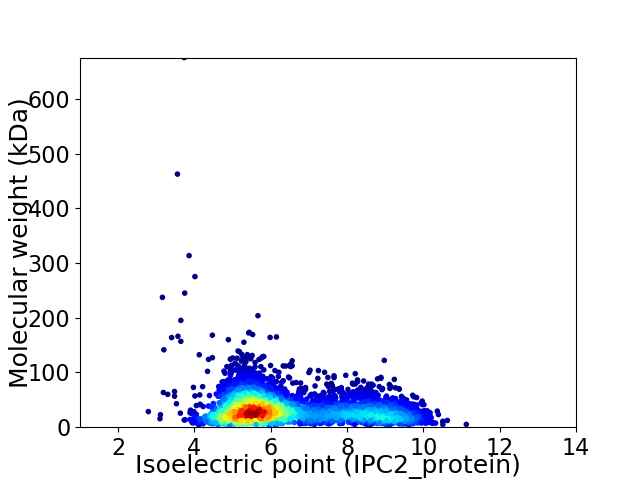
Virtual 2D-PAGE plot for 3840 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
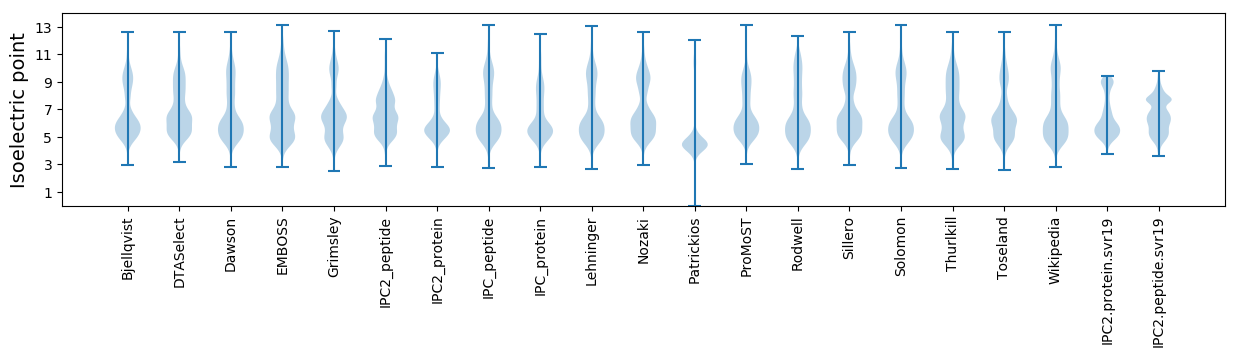
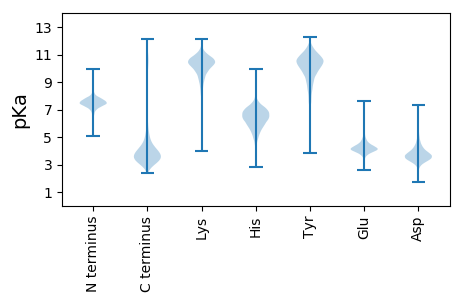
Protein with the lowest isoelectric point:
>tr|A0A7C9UVQ7|A0A7C9UVQ7_9PROT FeoC domain-containing protein OS=Magnetospirillum aberrantis SpK OX=908842 GN=G4223_05660 PE=4 SV=1
MM1 pKa = 7.07TLSSAMTTAVSALKK15 pKa = 10.36AQSQALSAISNNLANSSTTGYY36 pKa = 10.93KK37 pKa = 10.14SVTTSFQSLVTQAFSGTAYY56 pKa = 9.32TGAGVTSSVRR66 pKa = 11.84QNVDD70 pKa = 2.57AQGNIEE76 pKa = 4.09ATSNVTDD83 pKa = 3.76MAISGDD89 pKa = 3.46GMFVVTDD96 pKa = 3.45SSGAQYY102 pKa = 8.15YY103 pKa = 8.18TRR105 pKa = 11.84NGEE108 pKa = 3.9FDD110 pKa = 3.56VDD112 pKa = 4.06DD113 pKa = 6.39DD114 pKa = 4.94GYY116 pKa = 11.42LYY118 pKa = 11.32NNASTKK124 pKa = 10.35VYY126 pKa = 10.85LMGYY130 pKa = 9.65AINSDD135 pKa = 2.99GTTAKK140 pKa = 9.63TLSQINVTDD149 pKa = 4.04QDD151 pKa = 4.27TSWKK155 pKa = 7.72EE156 pKa = 4.0TTAVSLDD163 pKa = 3.37ASLGADD169 pKa = 3.34TDD171 pKa = 4.18VSSSTTYY178 pKa = 10.23EE179 pKa = 3.96KK180 pKa = 10.4TFEE183 pKa = 4.7VYY185 pKa = 10.82DD186 pKa = 3.84SMGGSHH192 pKa = 7.09IVTATFAHH200 pKa = 5.84TAVDD204 pKa = 3.94SSANTSTWTVSFSSDD219 pKa = 4.44DD220 pKa = 3.49GTTTLDD226 pKa = 3.32TLTLEE231 pKa = 4.43FDD233 pKa = 3.77GDD235 pKa = 4.23GNLTSATDD243 pKa = 3.61SSNVTVDD250 pKa = 3.79PSSLTLDD257 pKa = 4.0FDD259 pKa = 4.34WNNASSDD266 pKa = 3.57SSITLDD272 pKa = 3.75LSSISMGGSTDD283 pKa = 3.64GVTLNDD289 pKa = 3.64VTNDD293 pKa = 3.35GYY295 pKa = 11.52EE296 pKa = 3.94LGNLSGITISSDD308 pKa = 3.23GTVSASYY315 pKa = 11.43DD316 pKa = 3.37NGQTISLYY324 pKa = 9.98KK325 pKa = 10.27VAVATFANYY334 pKa = 10.21DD335 pKa = 3.47GLAALSGTLYY345 pKa = 10.54QQTADD350 pKa = 3.56SGEE353 pKa = 4.39AIIGTAEE360 pKa = 3.95TGSAGSISASSLEE373 pKa = 4.16SSTVDD378 pKa = 2.94TADD381 pKa = 2.92EE382 pKa = 4.26FTRR385 pKa = 11.84MIVAQQAYY393 pKa = 9.14SAASQVITTAKK404 pKa = 10.62DD405 pKa = 3.01MYY407 pKa = 10.46DD408 pKa = 3.38ALISAVRR415 pKa = 3.45
MM1 pKa = 7.07TLSSAMTTAVSALKK15 pKa = 10.36AQSQALSAISNNLANSSTTGYY36 pKa = 10.93KK37 pKa = 10.14SVTTSFQSLVTQAFSGTAYY56 pKa = 9.32TGAGVTSSVRR66 pKa = 11.84QNVDD70 pKa = 2.57AQGNIEE76 pKa = 4.09ATSNVTDD83 pKa = 3.76MAISGDD89 pKa = 3.46GMFVVTDD96 pKa = 3.45SSGAQYY102 pKa = 8.15YY103 pKa = 8.18TRR105 pKa = 11.84NGEE108 pKa = 3.9FDD110 pKa = 3.56VDD112 pKa = 4.06DD113 pKa = 6.39DD114 pKa = 4.94GYY116 pKa = 11.42LYY118 pKa = 11.32NNASTKK124 pKa = 10.35VYY126 pKa = 10.85LMGYY130 pKa = 9.65AINSDD135 pKa = 2.99GTTAKK140 pKa = 9.63TLSQINVTDD149 pKa = 4.04QDD151 pKa = 4.27TSWKK155 pKa = 7.72EE156 pKa = 4.0TTAVSLDD163 pKa = 3.37ASLGADD169 pKa = 3.34TDD171 pKa = 4.18VSSSTTYY178 pKa = 10.23EE179 pKa = 3.96KK180 pKa = 10.4TFEE183 pKa = 4.7VYY185 pKa = 10.82DD186 pKa = 3.84SMGGSHH192 pKa = 7.09IVTATFAHH200 pKa = 5.84TAVDD204 pKa = 3.94SSANTSTWTVSFSSDD219 pKa = 4.44DD220 pKa = 3.49GTTTLDD226 pKa = 3.32TLTLEE231 pKa = 4.43FDD233 pKa = 3.77GDD235 pKa = 4.23GNLTSATDD243 pKa = 3.61SSNVTVDD250 pKa = 3.79PSSLTLDD257 pKa = 4.0FDD259 pKa = 4.34WNNASSDD266 pKa = 3.57SSITLDD272 pKa = 3.75LSSISMGGSTDD283 pKa = 3.64GVTLNDD289 pKa = 3.64VTNDD293 pKa = 3.35GYY295 pKa = 11.52EE296 pKa = 3.94LGNLSGITISSDD308 pKa = 3.23GTVSASYY315 pKa = 11.43DD316 pKa = 3.37NGQTISLYY324 pKa = 9.98KK325 pKa = 10.27VAVATFANYY334 pKa = 10.21DD335 pKa = 3.47GLAALSGTLYY345 pKa = 10.54QQTADD350 pKa = 3.56SGEE353 pKa = 4.39AIIGTAEE360 pKa = 3.95TGSAGSISASSLEE373 pKa = 4.16SSTVDD378 pKa = 2.94TADD381 pKa = 2.92EE382 pKa = 4.26FTRR385 pKa = 11.84MIVAQQAYY393 pKa = 9.14SAASQVITTAKK404 pKa = 10.62DD405 pKa = 3.01MYY407 pKa = 10.46DD408 pKa = 3.38ALISAVRR415 pKa = 3.45
Molecular weight: 42.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7C9QRW5|A0A7C9QRW5_9PROT DUF2927 domain-containing protein OS=Magnetospirillum aberrantis SpK OX=908842 GN=G4223_01420 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.33VIATRR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.33VIATRR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1252964 |
29 |
6672 |
326.3 |
35.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.765 ± 0.058 | 0.943 ± 0.016 |
5.965 ± 0.039 | 5.714 ± 0.042 |
3.404 ± 0.026 | 8.552 ± 0.056 |
2.137 ± 0.024 | 4.294 ± 0.029 |
3.205 ± 0.031 | 10.49 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.545 ± 0.02 | 2.476 ± 0.028 |
5.094 ± 0.046 | 3.176 ± 0.024 |
7.215 ± 0.06 | 5.144 ± 0.042 |
5.32 ± 0.061 | 8.189 ± 0.037 |
1.353 ± 0.016 | 2.018 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
