
Korarchaeum cryptofilum (strain OPF8)
Taxonomy: cellular organisms; Archaea; TACK group; Candidatus Korarchaeota; Candidatus Korarchaeum; Candidatus Korarchaeum cryptofilum
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
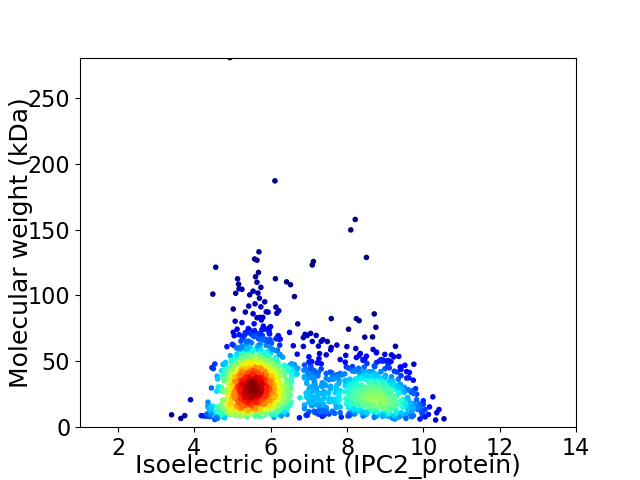
Virtual 2D-PAGE plot for 1602 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
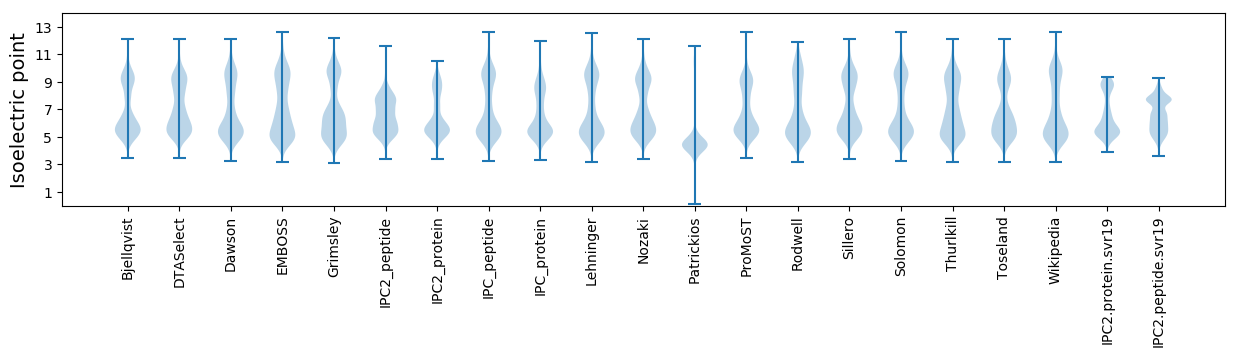
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B1L530|B1L530_KORCO Lysine biosynthesis enzyme LysX OS=Korarchaeum cryptofilum (strain OPF8) OX=374847 GN=Kcr_0812 PE=3 SV=1
MM1 pKa = 7.04FASLGANMEE10 pKa = 4.3ARR12 pKa = 11.84CPICGSTLEE21 pKa = 4.57VPDD24 pKa = 4.29DD25 pKa = 5.01AIPGEE30 pKa = 4.87LIDD33 pKa = 5.22CNSCGALLEE42 pKa = 4.26VFNDD46 pKa = 3.18NGAISLRR53 pKa = 11.84EE54 pKa = 3.88AGGVLEE60 pKa = 5.05DD61 pKa = 3.44WGEE64 pKa = 3.73
MM1 pKa = 7.04FASLGANMEE10 pKa = 4.3ARR12 pKa = 11.84CPICGSTLEE21 pKa = 4.57VPDD24 pKa = 4.29DD25 pKa = 5.01AIPGEE30 pKa = 4.87LIDD33 pKa = 5.22CNSCGALLEE42 pKa = 4.26VFNDD46 pKa = 3.18NGAISLRR53 pKa = 11.84EE54 pKa = 3.88AGGVLEE60 pKa = 5.05DD61 pKa = 3.44WGEE64 pKa = 3.73
Molecular weight: 6.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|B1L6Q8|GGGPS_KORCO Geranylgeranylglyceryl phosphate synthase OS=Korarchaeum cryptofilum (strain OPF8) OX=374847 GN=Kcr_1391 PE=3 SV=2
MM1 pKa = 8.29LEE3 pKa = 3.9IFRR6 pKa = 11.84TGQLEE11 pKa = 3.98SSYY14 pKa = 11.61ARR16 pKa = 11.84LNPLVKK22 pKa = 10.49FILFITFIIIPLISTSFLLQLFSLVAQIPLILMSRR57 pKa = 11.84SGRR60 pKa = 11.84RR61 pKa = 11.84VVRR64 pKa = 11.84SLKK67 pKa = 10.65ASTFFILILIVLNYY81 pKa = 8.34ITTNSIVFSLSMVIRR96 pKa = 11.84LLTMIIASAIFMNGSNPSEE115 pKa = 4.2IGDD118 pKa = 3.79VLSKK122 pKa = 10.91LRR124 pKa = 11.84VPTSITFSFIVALRR138 pKa = 11.84FIPVLADD145 pKa = 3.24DD146 pKa = 4.97FMNIIASQASRR157 pKa = 11.84GHH159 pKa = 4.62EE160 pKa = 3.93VEE162 pKa = 3.49RR163 pKa = 11.84SGFIRR168 pKa = 11.84RR169 pKa = 11.84AKK171 pKa = 10.5SLLPLLIPLIVIAIRR186 pKa = 11.84RR187 pKa = 11.84AQQLAEE193 pKa = 4.12ALEE196 pKa = 4.41SRR198 pKa = 11.84CFGSGKK204 pKa = 7.44RR205 pKa = 11.84TSYY208 pKa = 8.77ITYY211 pKa = 10.42EE212 pKa = 3.68MGLIDD217 pKa = 6.11FLALVYY223 pKa = 9.35ATAIMITGSLLATLPPEE240 pKa = 4.17FLLLPFRR247 pKa = 4.93
MM1 pKa = 8.29LEE3 pKa = 3.9IFRR6 pKa = 11.84TGQLEE11 pKa = 3.98SSYY14 pKa = 11.61ARR16 pKa = 11.84LNPLVKK22 pKa = 10.49FILFITFIIIPLISTSFLLQLFSLVAQIPLILMSRR57 pKa = 11.84SGRR60 pKa = 11.84RR61 pKa = 11.84VVRR64 pKa = 11.84SLKK67 pKa = 10.65ASTFFILILIVLNYY81 pKa = 8.34ITTNSIVFSLSMVIRR96 pKa = 11.84LLTMIIASAIFMNGSNPSEE115 pKa = 4.2IGDD118 pKa = 3.79VLSKK122 pKa = 10.91LRR124 pKa = 11.84VPTSITFSFIVALRR138 pKa = 11.84FIPVLADD145 pKa = 3.24DD146 pKa = 4.97FMNIIASQASRR157 pKa = 11.84GHH159 pKa = 4.62EE160 pKa = 3.93VEE162 pKa = 3.49RR163 pKa = 11.84SGFIRR168 pKa = 11.84RR169 pKa = 11.84AKK171 pKa = 10.5SLLPLLIPLIVIAIRR186 pKa = 11.84RR187 pKa = 11.84AQQLAEE193 pKa = 4.12ALEE196 pKa = 4.41SRR198 pKa = 11.84CFGSGKK204 pKa = 7.44RR205 pKa = 11.84TSYY208 pKa = 8.77ITYY211 pKa = 10.42EE212 pKa = 3.68MGLIDD217 pKa = 6.11FLALVYY223 pKa = 9.35ATAIMITGSLLATLPPEE240 pKa = 4.17FLLLPFRR247 pKa = 4.93
Molecular weight: 27.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
475001 |
47 |
2546 |
296.5 |
33.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.974 ± 0.066 | 0.845 ± 0.025 |
4.932 ± 0.037 | 8.157 ± 0.078 |
3.84 ± 0.037 | 7.787 ± 0.049 |
1.368 ± 0.021 | 8.199 ± 0.048 |
5.564 ± 0.05 | 10.877 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.656 ± 0.026 | 2.822 ± 0.037 |
4.488 ± 0.039 | 1.532 ± 0.024 |
6.994 ± 0.064 | 7.328 ± 0.069 |
3.742 ± 0.045 | 7.294 ± 0.043 |
1.136 ± 0.021 | 3.465 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
