
Hubei sobemo-like virus 41
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.6
Get precalculated fractions of proteins
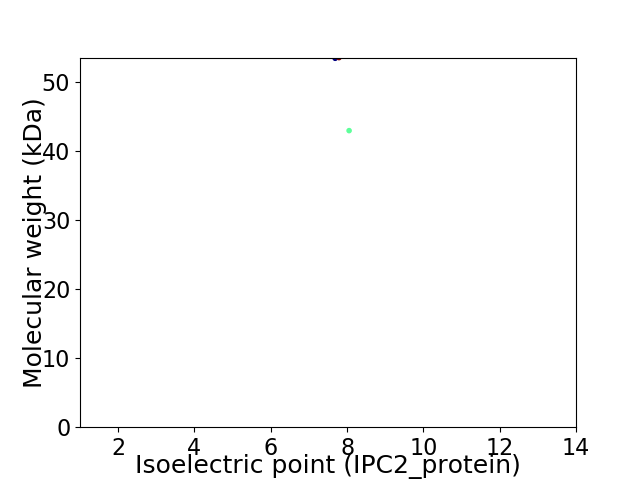
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE25|A0A1L3KE25_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 41 OX=1923229 PE=4 SV=1
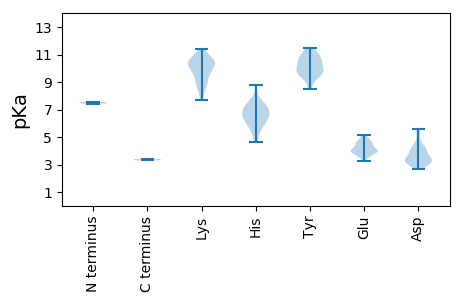
MM1 pKa = 7.55YY2 pKa = 10.39IPVNKK7 pKa = 9.66VLYY10 pKa = 10.54VDD12 pKa = 4.37VIEE15 pKa = 5.11LKK17 pKa = 10.48IRR19 pKa = 11.84DD20 pKa = 3.27TWAAIEE26 pKa = 4.57SYY28 pKa = 11.44LMDD31 pKa = 3.5TPYY34 pKa = 10.84CRR36 pKa = 11.84TVEE39 pKa = 3.99SGFWFWKK46 pKa = 7.73EE47 pKa = 3.75THH49 pKa = 6.72CEE51 pKa = 4.06LRR53 pKa = 11.84WVPLMVTILLALVSYY68 pKa = 10.15FGLKK72 pKa = 10.15RR73 pKa = 11.84IVRR76 pKa = 11.84GCLKK80 pKa = 9.53PRR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84VTYY87 pKa = 9.89EE88 pKa = 3.46VDD90 pKa = 3.25PLLTQQEE97 pKa = 4.62SLVIGKK103 pKa = 9.06PLMPGGKK110 pKa = 8.71QPKK113 pKa = 9.23CQVTLATRR121 pKa = 11.84RR122 pKa = 11.84NDD124 pKa = 2.88VRR126 pKa = 11.84LLMGGGFRR134 pKa = 11.84LLDD137 pKa = 3.65YY138 pKa = 10.85LVLPTHH144 pKa = 6.72VMHH147 pKa = 7.33AGQGKK152 pKa = 9.92VYY154 pKa = 10.87VLANDD159 pKa = 4.15KK160 pKa = 10.52EE161 pKa = 4.32LEE163 pKa = 4.11IPKK166 pKa = 10.22DD167 pKa = 3.41AEE169 pKa = 4.53TIDD172 pKa = 4.05LAADD176 pKa = 4.18LLAVKK181 pKa = 10.29ISQAEE186 pKa = 3.86WSKK189 pKa = 11.11IGVSQAKK196 pKa = 9.29LGPMSGTPTVQVTSPCDD213 pKa = 3.19QKK215 pKa = 11.43FSIGYY220 pKa = 9.39LKK222 pKa = 10.4LAKK225 pKa = 10.37SFGRR229 pKa = 11.84VEE231 pKa = 4.44YY232 pKa = 10.53DD233 pKa = 3.11SSTQPGFSGAAYY245 pKa = 9.87MNGNVCLGMHH255 pKa = 6.31LHH257 pKa = 6.51GGVVAGGYY265 pKa = 9.22EE266 pKa = 3.89GLYY269 pKa = 9.68IYY271 pKa = 11.01ARR273 pKa = 11.84LKK275 pKa = 9.55HH276 pKa = 5.94HH277 pKa = 7.26LYY279 pKa = 9.88MNGVEE284 pKa = 5.0DD285 pKa = 4.35EE286 pKa = 4.4ASKK289 pKa = 9.93TSPGGSDD296 pKa = 3.19WAPDD300 pKa = 3.32YY301 pKa = 11.43HH302 pKa = 7.48DD303 pKa = 3.64INEE306 pKa = 4.13FEE308 pKa = 4.39EE309 pKa = 4.79LEE311 pKa = 4.08QKK313 pKa = 10.35IVDD316 pKa = 3.94GKK318 pKa = 8.78SEE320 pKa = 3.6RR321 pKa = 11.84RR322 pKa = 11.84AIVRR326 pKa = 11.84LKK328 pKa = 10.19SGHH331 pKa = 4.62YY332 pKa = 9.92HH333 pKa = 5.22ITRR336 pKa = 11.84GEE338 pKa = 4.06LLDD341 pKa = 4.24KK342 pKa = 10.74INRR345 pKa = 11.84LRR347 pKa = 11.84DD348 pKa = 3.41SNAWADD354 pKa = 3.46QMEE357 pKa = 4.66AEE359 pKa = 4.73DD360 pKa = 5.35AQAQLDD366 pKa = 3.88NKK368 pKa = 10.52EE369 pKa = 4.63YY370 pKa = 10.19IPEE373 pKa = 4.17AAIHH377 pKa = 6.08PGKK380 pKa = 10.48GVEE383 pKa = 4.64FSGEE387 pKa = 4.12GQRR390 pKa = 11.84PAGRR394 pKa = 11.84AAPGQEE400 pKa = 3.85QSRR403 pKa = 11.84SHH405 pKa = 6.79SDD407 pKa = 3.02SKK409 pKa = 11.4SSNGEE414 pKa = 3.27PRR416 pKa = 11.84RR417 pKa = 11.84PRR419 pKa = 11.84TEE421 pKa = 3.61RR422 pKa = 11.84DD423 pKa = 3.16RR424 pKa = 11.84LLLEE428 pKa = 4.53LSSVSNLQLRR438 pKa = 11.84EE439 pKa = 3.84YY440 pKa = 9.25LTSVKK445 pKa = 10.3NGRR448 pKa = 11.84RR449 pKa = 11.84QARR452 pKa = 11.84AMSLQIPPQEE462 pKa = 4.16RR463 pKa = 11.84NGNRR467 pKa = 11.84SALNSTEE474 pKa = 3.82SRR476 pKa = 11.84VV477 pKa = 3.31
MM1 pKa = 7.55YY2 pKa = 10.39IPVNKK7 pKa = 9.66VLYY10 pKa = 10.54VDD12 pKa = 4.37VIEE15 pKa = 5.11LKK17 pKa = 10.48IRR19 pKa = 11.84DD20 pKa = 3.27TWAAIEE26 pKa = 4.57SYY28 pKa = 11.44LMDD31 pKa = 3.5TPYY34 pKa = 10.84CRR36 pKa = 11.84TVEE39 pKa = 3.99SGFWFWKK46 pKa = 7.73EE47 pKa = 3.75THH49 pKa = 6.72CEE51 pKa = 4.06LRR53 pKa = 11.84WVPLMVTILLALVSYY68 pKa = 10.15FGLKK72 pKa = 10.15RR73 pKa = 11.84IVRR76 pKa = 11.84GCLKK80 pKa = 9.53PRR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84VTYY87 pKa = 9.89EE88 pKa = 3.46VDD90 pKa = 3.25PLLTQQEE97 pKa = 4.62SLVIGKK103 pKa = 9.06PLMPGGKK110 pKa = 8.71QPKK113 pKa = 9.23CQVTLATRR121 pKa = 11.84RR122 pKa = 11.84NDD124 pKa = 2.88VRR126 pKa = 11.84LLMGGGFRR134 pKa = 11.84LLDD137 pKa = 3.65YY138 pKa = 10.85LVLPTHH144 pKa = 6.72VMHH147 pKa = 7.33AGQGKK152 pKa = 9.92VYY154 pKa = 10.87VLANDD159 pKa = 4.15KK160 pKa = 10.52EE161 pKa = 4.32LEE163 pKa = 4.11IPKK166 pKa = 10.22DD167 pKa = 3.41AEE169 pKa = 4.53TIDD172 pKa = 4.05LAADD176 pKa = 4.18LLAVKK181 pKa = 10.29ISQAEE186 pKa = 3.86WSKK189 pKa = 11.11IGVSQAKK196 pKa = 9.29LGPMSGTPTVQVTSPCDD213 pKa = 3.19QKK215 pKa = 11.43FSIGYY220 pKa = 9.39LKK222 pKa = 10.4LAKK225 pKa = 10.37SFGRR229 pKa = 11.84VEE231 pKa = 4.44YY232 pKa = 10.53DD233 pKa = 3.11SSTQPGFSGAAYY245 pKa = 9.87MNGNVCLGMHH255 pKa = 6.31LHH257 pKa = 6.51GGVVAGGYY265 pKa = 9.22EE266 pKa = 3.89GLYY269 pKa = 9.68IYY271 pKa = 11.01ARR273 pKa = 11.84LKK275 pKa = 9.55HH276 pKa = 5.94HH277 pKa = 7.26LYY279 pKa = 9.88MNGVEE284 pKa = 5.0DD285 pKa = 4.35EE286 pKa = 4.4ASKK289 pKa = 9.93TSPGGSDD296 pKa = 3.19WAPDD300 pKa = 3.32YY301 pKa = 11.43HH302 pKa = 7.48DD303 pKa = 3.64INEE306 pKa = 4.13FEE308 pKa = 4.39EE309 pKa = 4.79LEE311 pKa = 4.08QKK313 pKa = 10.35IVDD316 pKa = 3.94GKK318 pKa = 8.78SEE320 pKa = 3.6RR321 pKa = 11.84RR322 pKa = 11.84AIVRR326 pKa = 11.84LKK328 pKa = 10.19SGHH331 pKa = 4.62YY332 pKa = 9.92HH333 pKa = 5.22ITRR336 pKa = 11.84GEE338 pKa = 4.06LLDD341 pKa = 4.24KK342 pKa = 10.74INRR345 pKa = 11.84LRR347 pKa = 11.84DD348 pKa = 3.41SNAWADD354 pKa = 3.46QMEE357 pKa = 4.66AEE359 pKa = 4.73DD360 pKa = 5.35AQAQLDD366 pKa = 3.88NKK368 pKa = 10.52EE369 pKa = 4.63YY370 pKa = 10.19IPEE373 pKa = 4.17AAIHH377 pKa = 6.08PGKK380 pKa = 10.48GVEE383 pKa = 4.64FSGEE387 pKa = 4.12GQRR390 pKa = 11.84PAGRR394 pKa = 11.84AAPGQEE400 pKa = 3.85QSRR403 pKa = 11.84SHH405 pKa = 6.79SDD407 pKa = 3.02SKK409 pKa = 11.4SSNGEE414 pKa = 3.27PRR416 pKa = 11.84RR417 pKa = 11.84PRR419 pKa = 11.84TEE421 pKa = 3.61RR422 pKa = 11.84DD423 pKa = 3.16RR424 pKa = 11.84LLLEE428 pKa = 4.53LSSVSNLQLRR438 pKa = 11.84EE439 pKa = 3.84YY440 pKa = 9.25LTSVKK445 pKa = 10.3NGRR448 pKa = 11.84RR449 pKa = 11.84QARR452 pKa = 11.84AMSLQIPPQEE462 pKa = 4.16RR463 pKa = 11.84NGNRR467 pKa = 11.84SALNSTEE474 pKa = 3.82SRR476 pKa = 11.84VV477 pKa = 3.31
Molecular weight: 53.4 kDa
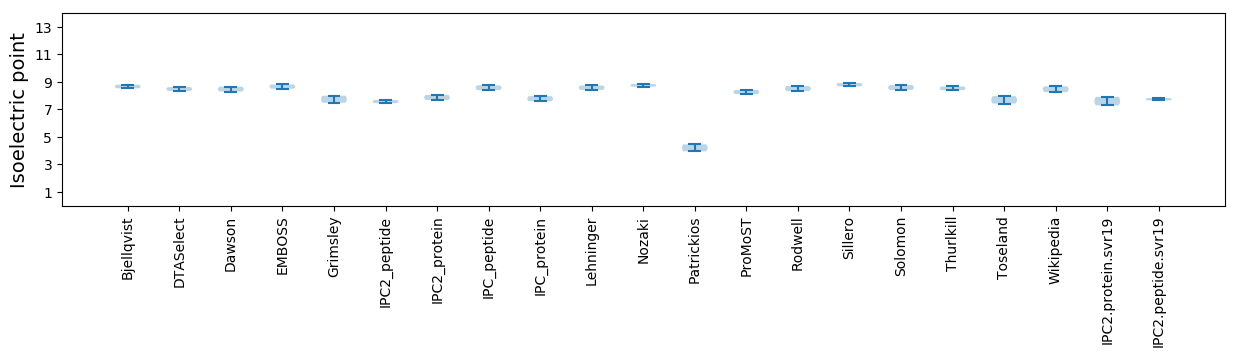
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE25|A0A1L3KE25_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 41 OX=1923229 PE=4 SV=1
MM1 pKa = 7.43HH2 pKa = 7.23SGYY5 pKa = 10.67VIPADD10 pKa = 3.51WNSKK14 pKa = 8.77SRR16 pKa = 11.84FMTLLLTLDD25 pKa = 4.09YY26 pKa = 10.39SSSPGYY32 pKa = 9.69PYY34 pKa = 10.73LRR36 pKa = 11.84EE37 pKa = 4.07APTIGKK43 pKa = 8.3WLRR46 pKa = 11.84TDD48 pKa = 3.06EE49 pKa = 4.99CGSFDD54 pKa = 3.7PVQVEE59 pKa = 4.13RR60 pKa = 11.84LWYY63 pKa = 9.39DD64 pKa = 3.08VNMVMAGSYY73 pKa = 8.52EE74 pKa = 3.96HH75 pKa = 7.5LFRR78 pKa = 11.84AFVKK82 pKa = 10.68DD83 pKa = 4.06EE84 pKa = 3.7PHH86 pKa = 6.91KK87 pKa = 10.32IAKK90 pKa = 9.95AKK92 pKa = 8.56EE93 pKa = 3.88NRR95 pKa = 11.84WRR97 pKa = 11.84LIIASSLPVQMVWRR111 pKa = 11.84MLYY114 pKa = 9.65TGQNEE119 pKa = 4.52ALNKK123 pKa = 8.89YY124 pKa = 10.19HH125 pKa = 7.33DD126 pKa = 4.13WCPSKK131 pKa = 10.78HH132 pKa = 5.94GFVFCYY138 pKa = 9.65GGWMKK143 pKa = 10.43FIAQAKK149 pKa = 7.73TKK151 pKa = 9.54GLNVSRR157 pKa = 11.84DD158 pKa = 3.04ISGWDD163 pKa = 3.06VGAPGWVFEE172 pKa = 4.66VVGAWRR178 pKa = 11.84EE179 pKa = 3.94SWPRR183 pKa = 11.84ATDD186 pKa = 2.67SWIRR190 pKa = 11.84VHH192 pKa = 8.75RR193 pKa = 11.84MMYY196 pKa = 10.67DD197 pKa = 3.29DD198 pKa = 5.04AYY200 pKa = 9.67KK201 pKa = 10.16SSRR204 pKa = 11.84IIFSNGVVVRR214 pKa = 11.84QLFGGFMKK222 pKa = 10.58SGLFNTISDD231 pKa = 3.57NSLAMGAIHH240 pKa = 6.95SLACVRR246 pKa = 11.84SGLPFGHH253 pKa = 6.63YY254 pKa = 9.78IVTGDD259 pKa = 5.56DD260 pKa = 3.21IAQSTVSAGYY270 pKa = 10.76LDD272 pKa = 4.71ALYY275 pKa = 11.13KK276 pKa = 10.47LGCRR280 pKa = 11.84VKK282 pKa = 10.17EE283 pKa = 3.97VLYY286 pKa = 10.22HH287 pKa = 6.0LEE289 pKa = 3.86FMGVNFSSGKK299 pKa = 8.65PEE301 pKa = 3.65PMYY304 pKa = 9.36TQKK307 pKa = 11.01HH308 pKa = 5.49LFNLLTKK315 pKa = 9.53EE316 pKa = 3.92QFIAEE321 pKa = 4.38TMDD324 pKa = 3.28SYY326 pKa = 11.47CRR328 pKa = 11.84YY329 pKa = 8.72YY330 pKa = 11.12AEE332 pKa = 3.96SSKK335 pKa = 9.47YY336 pKa = 10.03HH337 pKa = 6.61WFTEE341 pKa = 4.14IAKK344 pKa = 9.8EE345 pKa = 3.56LAVPVKK351 pKa = 10.68SRR353 pKa = 11.84WFYY356 pKa = 9.4QFWYY360 pKa = 9.91SSPLAEE366 pKa = 5.03IYY368 pKa = 10.47HH369 pKa = 6.25KK370 pKa = 10.98LGG372 pKa = 3.41
MM1 pKa = 7.43HH2 pKa = 7.23SGYY5 pKa = 10.67VIPADD10 pKa = 3.51WNSKK14 pKa = 8.77SRR16 pKa = 11.84FMTLLLTLDD25 pKa = 4.09YY26 pKa = 10.39SSSPGYY32 pKa = 9.69PYY34 pKa = 10.73LRR36 pKa = 11.84EE37 pKa = 4.07APTIGKK43 pKa = 8.3WLRR46 pKa = 11.84TDD48 pKa = 3.06EE49 pKa = 4.99CGSFDD54 pKa = 3.7PVQVEE59 pKa = 4.13RR60 pKa = 11.84LWYY63 pKa = 9.39DD64 pKa = 3.08VNMVMAGSYY73 pKa = 8.52EE74 pKa = 3.96HH75 pKa = 7.5LFRR78 pKa = 11.84AFVKK82 pKa = 10.68DD83 pKa = 4.06EE84 pKa = 3.7PHH86 pKa = 6.91KK87 pKa = 10.32IAKK90 pKa = 9.95AKK92 pKa = 8.56EE93 pKa = 3.88NRR95 pKa = 11.84WRR97 pKa = 11.84LIIASSLPVQMVWRR111 pKa = 11.84MLYY114 pKa = 9.65TGQNEE119 pKa = 4.52ALNKK123 pKa = 8.89YY124 pKa = 10.19HH125 pKa = 7.33DD126 pKa = 4.13WCPSKK131 pKa = 10.78HH132 pKa = 5.94GFVFCYY138 pKa = 9.65GGWMKK143 pKa = 10.43FIAQAKK149 pKa = 7.73TKK151 pKa = 9.54GLNVSRR157 pKa = 11.84DD158 pKa = 3.04ISGWDD163 pKa = 3.06VGAPGWVFEE172 pKa = 4.66VVGAWRR178 pKa = 11.84EE179 pKa = 3.94SWPRR183 pKa = 11.84ATDD186 pKa = 2.67SWIRR190 pKa = 11.84VHH192 pKa = 8.75RR193 pKa = 11.84MMYY196 pKa = 10.67DD197 pKa = 3.29DD198 pKa = 5.04AYY200 pKa = 9.67KK201 pKa = 10.16SSRR204 pKa = 11.84IIFSNGVVVRR214 pKa = 11.84QLFGGFMKK222 pKa = 10.58SGLFNTISDD231 pKa = 3.57NSLAMGAIHH240 pKa = 6.95SLACVRR246 pKa = 11.84SGLPFGHH253 pKa = 6.63YY254 pKa = 9.78IVTGDD259 pKa = 5.56DD260 pKa = 3.21IAQSTVSAGYY270 pKa = 10.76LDD272 pKa = 4.71ALYY275 pKa = 11.13KK276 pKa = 10.47LGCRR280 pKa = 11.84VKK282 pKa = 10.17EE283 pKa = 3.97VLYY286 pKa = 10.22HH287 pKa = 6.0LEE289 pKa = 3.86FMGVNFSSGKK299 pKa = 8.65PEE301 pKa = 3.65PMYY304 pKa = 9.36TQKK307 pKa = 11.01HH308 pKa = 5.49LFNLLTKK315 pKa = 9.53EE316 pKa = 3.92QFIAEE321 pKa = 4.38TMDD324 pKa = 3.28SYY326 pKa = 11.47CRR328 pKa = 11.84YY329 pKa = 8.72YY330 pKa = 11.12AEE332 pKa = 3.96SSKK335 pKa = 9.47YY336 pKa = 10.03HH337 pKa = 6.61WFTEE341 pKa = 4.14IAKK344 pKa = 9.8EE345 pKa = 3.56LAVPVKK351 pKa = 10.68SRR353 pKa = 11.84WFYY356 pKa = 9.4QFWYY360 pKa = 9.91SSPLAEE366 pKa = 5.03IYY368 pKa = 10.47HH369 pKa = 6.25KK370 pKa = 10.98LGG372 pKa = 3.41
Molecular weight: 42.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
849 |
372 |
477 |
424.5 |
48.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.832 ± 0.077 | 1.413 ± 0.138 |
4.829 ± 0.18 | 6.125 ± 0.891 |
3.416 ± 1.358 | 7.892 ± 0.439 |
2.827 ± 0.276 | 4.476 ± 0.065 |
5.771 ± 0.099 | 9.187 ± 0.964 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.062 ± 0.486 | 3.18 ± 0.155 |
4.594 ± 0.389 | 3.534 ± 0.772 |
6.36 ± 0.868 | 8.009 ± 0.597 |
4.005 ± 0.167 | 6.949 ± 0.028 |
2.591 ± 0.998 | 4.947 ± 0.856 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
