
Pelagibacter phage HTVC200P
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Votkovvirus; unclassified Votkovvirus
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
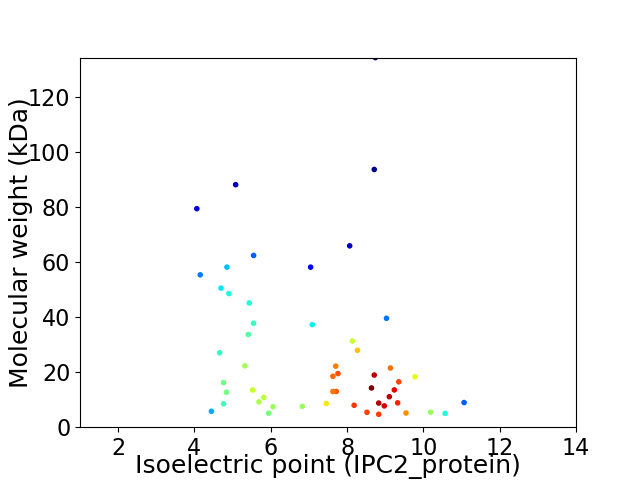
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y1NTV0|A0A4Y1NTV0_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC200P OX=2283023 GN=P200_gp041 PE=4 SV=1
MM1 pKa = 7.57ANSFLEE7 pKa = 4.17YY8 pKa = 9.72TGNGSTTAFSITFDD22 pKa = 3.78YY23 pKa = 11.26LDD25 pKa = 4.44ASHH28 pKa = 7.37IACTVDD34 pKa = 3.62GVSTSFTLSNGGTTATLAAAPANGAAIRR62 pKa = 11.84FTRR65 pKa = 11.84TTSQATRR72 pKa = 11.84LTDD75 pKa = 3.51YY76 pKa = 10.99VAGSVLTEE84 pKa = 3.99TDD86 pKa = 3.77LDD88 pKa = 4.05TDD90 pKa = 3.98SKK92 pKa = 11.27QAFFMGQEE100 pKa = 4.19GLDD103 pKa = 3.76TIGTKK108 pKa = 9.4MGQSISTFQFDD119 pKa = 4.16ALNKK123 pKa = 10.07RR124 pKa = 11.84ITNVADD130 pKa = 3.26PTSAQDD136 pKa = 3.52AATKK140 pKa = 10.59NYY142 pKa = 10.63LEE144 pKa = 4.22NTWLSSTDD152 pKa = 3.32KK153 pKa = 10.82ATLNSVNANIANINAVNSNSSNINSAVSNATNINTVATNIGSVNTVAADD202 pKa = 3.23ITKK205 pKa = 9.97VVAVANDD212 pKa = 3.39LAEE215 pKa = 4.27AVSEE219 pKa = 4.51VEE221 pKa = 4.71TVADD225 pKa = 4.55DD226 pKa = 4.36LNEE229 pKa = 4.13ATSEE233 pKa = 3.84IDD235 pKa = 3.32TVANAITNVDD245 pKa = 3.25NVGNNIANVNSVASNATNINAVAGNATNINAVNSNSANINTVAGQNANITTLAGINANITTVAGIAANVTSVAGNEE321 pKa = 4.16TNINAVNSNSANINTVAGANSNITSVAGSIANVNTVATNLASVNSFANTYY371 pKa = 9.44RR372 pKa = 11.84ISSSAPTTSLDD383 pKa = 3.77VGDD386 pKa = 5.38LYY388 pKa = 11.5FDD390 pKa = 3.53TTANEE395 pKa = 3.92LKK397 pKa = 10.5VYY399 pKa = 10.44KK400 pKa = 10.46SSGWAAAGSTVNGTAQRR417 pKa = 11.84YY418 pKa = 5.38TYY420 pKa = 10.27NITGTPTTVSGSDD433 pKa = 3.32ANGNTLAYY441 pKa = 9.86DD442 pKa = 3.68AGYY445 pKa = 11.18ADD447 pKa = 4.47VYY449 pKa = 11.31LNGIRR454 pKa = 11.84MSSADD459 pKa = 3.2ITITSGTSVVFASALANGDD478 pKa = 3.63VVDD481 pKa = 4.41VVAYY485 pKa = 7.85GTFNVASINAANIDD499 pKa = 3.72AGTLNNARR507 pKa = 11.84LPSTISDD514 pKa = 3.45KK515 pKa = 10.93TIEE518 pKa = 4.22ATALTAKK525 pKa = 10.53GDD527 pKa = 3.63GSSADD532 pKa = 3.46GKK534 pKa = 9.41ITLNCSQNSHH544 pKa = 5.34GVAIQSPAHH553 pKa = 6.14SAGQSYY559 pKa = 7.71TLILPTSVGSNGQVLATAGSNTNQLTWVDD588 pKa = 3.61AQEE591 pKa = 4.39TKK593 pKa = 9.39PTVANVSQTIAPATATTITITGTNFVSIPHH623 pKa = 5.59VEE625 pKa = 4.27FVKK628 pKa = 10.31TDD630 pKa = 3.49GSVTIANTVSFTSSTSLSVNVTLASGNYY658 pKa = 7.4YY659 pKa = 10.35VRR661 pKa = 11.84IEE663 pKa = 4.31NPDD666 pKa = 3.26GNAGRR671 pKa = 11.84STQNIITASTAPSFTTSAGSLGTIAGNFSGTVATIAGTSDD711 pKa = 3.16SAITFSEE718 pKa = 4.55TTSVLTTANVTLNTSTGALTTTDD741 pKa = 4.03FGGSSTTPTTYY752 pKa = 11.06NFTIRR757 pKa = 11.84ITDD760 pKa = 3.64AEE762 pKa = 4.46GQTADD767 pKa = 4.08RR768 pKa = 11.84NFSLTSSFGATGGGQFNN785 pKa = 4.04
MM1 pKa = 7.57ANSFLEE7 pKa = 4.17YY8 pKa = 9.72TGNGSTTAFSITFDD22 pKa = 3.78YY23 pKa = 11.26LDD25 pKa = 4.44ASHH28 pKa = 7.37IACTVDD34 pKa = 3.62GVSTSFTLSNGGTTATLAAAPANGAAIRR62 pKa = 11.84FTRR65 pKa = 11.84TTSQATRR72 pKa = 11.84LTDD75 pKa = 3.51YY76 pKa = 10.99VAGSVLTEE84 pKa = 3.99TDD86 pKa = 3.77LDD88 pKa = 4.05TDD90 pKa = 3.98SKK92 pKa = 11.27QAFFMGQEE100 pKa = 4.19GLDD103 pKa = 3.76TIGTKK108 pKa = 9.4MGQSISTFQFDD119 pKa = 4.16ALNKK123 pKa = 10.07RR124 pKa = 11.84ITNVADD130 pKa = 3.26PTSAQDD136 pKa = 3.52AATKK140 pKa = 10.59NYY142 pKa = 10.63LEE144 pKa = 4.22NTWLSSTDD152 pKa = 3.32KK153 pKa = 10.82ATLNSVNANIANINAVNSNSSNINSAVSNATNINTVATNIGSVNTVAADD202 pKa = 3.23ITKK205 pKa = 9.97VVAVANDD212 pKa = 3.39LAEE215 pKa = 4.27AVSEE219 pKa = 4.51VEE221 pKa = 4.71TVADD225 pKa = 4.55DD226 pKa = 4.36LNEE229 pKa = 4.13ATSEE233 pKa = 3.84IDD235 pKa = 3.32TVANAITNVDD245 pKa = 3.25NVGNNIANVNSVASNATNINAVAGNATNINAVNSNSANINTVAGQNANITTLAGINANITTVAGIAANVTSVAGNEE321 pKa = 4.16TNINAVNSNSANINTVAGANSNITSVAGSIANVNTVATNLASVNSFANTYY371 pKa = 9.44RR372 pKa = 11.84ISSSAPTTSLDD383 pKa = 3.77VGDD386 pKa = 5.38LYY388 pKa = 11.5FDD390 pKa = 3.53TTANEE395 pKa = 3.92LKK397 pKa = 10.5VYY399 pKa = 10.44KK400 pKa = 10.46SSGWAAAGSTVNGTAQRR417 pKa = 11.84YY418 pKa = 5.38TYY420 pKa = 10.27NITGTPTTVSGSDD433 pKa = 3.32ANGNTLAYY441 pKa = 9.86DD442 pKa = 3.68AGYY445 pKa = 11.18ADD447 pKa = 4.47VYY449 pKa = 11.31LNGIRR454 pKa = 11.84MSSADD459 pKa = 3.2ITITSGTSVVFASALANGDD478 pKa = 3.63VVDD481 pKa = 4.41VVAYY485 pKa = 7.85GTFNVASINAANIDD499 pKa = 3.72AGTLNNARR507 pKa = 11.84LPSTISDD514 pKa = 3.45KK515 pKa = 10.93TIEE518 pKa = 4.22ATALTAKK525 pKa = 10.53GDD527 pKa = 3.63GSSADD532 pKa = 3.46GKK534 pKa = 9.41ITLNCSQNSHH544 pKa = 5.34GVAIQSPAHH553 pKa = 6.14SAGQSYY559 pKa = 7.71TLILPTSVGSNGQVLATAGSNTNQLTWVDD588 pKa = 3.61AQEE591 pKa = 4.39TKK593 pKa = 9.39PTVANVSQTIAPATATTITITGTNFVSIPHH623 pKa = 5.59VEE625 pKa = 4.27FVKK628 pKa = 10.31TDD630 pKa = 3.49GSVTIANTVSFTSSTSLSVNVTLASGNYY658 pKa = 7.4YY659 pKa = 10.35VRR661 pKa = 11.84IEE663 pKa = 4.31NPDD666 pKa = 3.26GNAGRR671 pKa = 11.84STQNIITASTAPSFTTSAGSLGTIAGNFSGTVATIAGTSDD711 pKa = 3.16SAITFSEE718 pKa = 4.55TTSVLTTANVTLNTSTGALTTTDD741 pKa = 4.03FGGSSTTPTTYY752 pKa = 11.06NFTIRR757 pKa = 11.84ITDD760 pKa = 3.64AEE762 pKa = 4.46GQTADD767 pKa = 4.08RR768 pKa = 11.84NFSLTSSFGATGGGQFNN785 pKa = 4.04
Molecular weight: 79.51 kDa
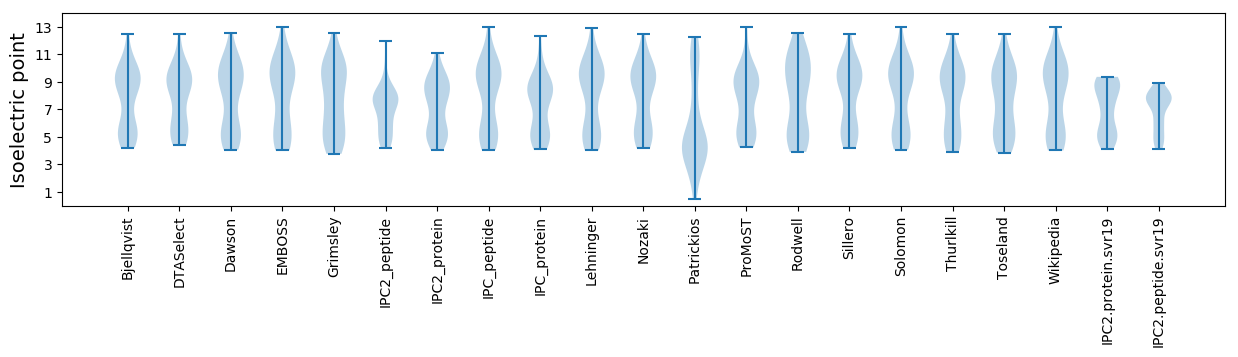
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y1NTT2|A0A4Y1NTT2_9CAUD Tail tubular protein B OS=Pelagibacter phage HTVC200P OX=2283023 GN=P200_gp029 PE=4 SV=1
MM1 pKa = 6.7VAKK4 pKa = 10.21RR5 pKa = 11.84YY6 pKa = 8.29QSPSGGLNARR16 pKa = 11.84GRR18 pKa = 11.84KK19 pKa = 9.07KK20 pKa = 10.61FGVKK24 pKa = 10.34APVKK28 pKa = 10.24KK29 pKa = 9.29GTNSRR34 pKa = 11.84RR35 pKa = 11.84VSFAARR41 pKa = 11.84FSGMKK46 pKa = 10.51GPMKK50 pKa = 10.65KK51 pKa = 9.93NGKK54 pKa = 5.21PTRR57 pKa = 11.84KK58 pKa = 9.84ALALKK63 pKa = 9.71KK64 pKa = 9.66WGFGSVAAARR74 pKa = 11.84KK75 pKa = 8.33FAAKK79 pKa = 9.83NKK81 pKa = 10.03KK82 pKa = 8.96SS83 pKa = 3.32
MM1 pKa = 6.7VAKK4 pKa = 10.21RR5 pKa = 11.84YY6 pKa = 8.29QSPSGGLNARR16 pKa = 11.84GRR18 pKa = 11.84KK19 pKa = 9.07KK20 pKa = 10.61FGVKK24 pKa = 10.34APVKK28 pKa = 10.24KK29 pKa = 9.29GTNSRR34 pKa = 11.84RR35 pKa = 11.84VSFAARR41 pKa = 11.84FSGMKK46 pKa = 10.51GPMKK50 pKa = 10.65KK51 pKa = 9.93NGKK54 pKa = 5.21PTRR57 pKa = 11.84KK58 pKa = 9.84ALALKK63 pKa = 9.71KK64 pKa = 9.66WGFGSVAAARR74 pKa = 11.84KK75 pKa = 8.33FAAKK79 pKa = 9.83NKK81 pKa = 10.03KK82 pKa = 8.96SS83 pKa = 3.32
Molecular weight: 8.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13289 |
40 |
1201 |
250.7 |
27.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.773 ± 0.467 | 0.903 ± 0.142 |
5.711 ± 0.234 | 5.584 ± 0.424 |
3.981 ± 0.185 | 7.668 ± 0.798 |
1.46 ± 0.199 | 6.569 ± 0.189 |
8.15 ± 0.729 | 7.668 ± 0.43 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.987 ± 0.165 | 6.532 ± 0.361 |
3.07 ± 0.232 | 3.544 ± 0.206 |
3.913 ± 0.297 | 7.442 ± 0.481 |
7.457 ± 0.674 | 5.599 ± 0.248 |
1.204 ± 0.148 | 3.785 ± 0.255 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
