
Halorubrum hochstenium ATCC 700873
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halorubrum; Halorubrum hochstenium
Average proteome isoelectric point is 4.82
Get precalculated fractions of proteins
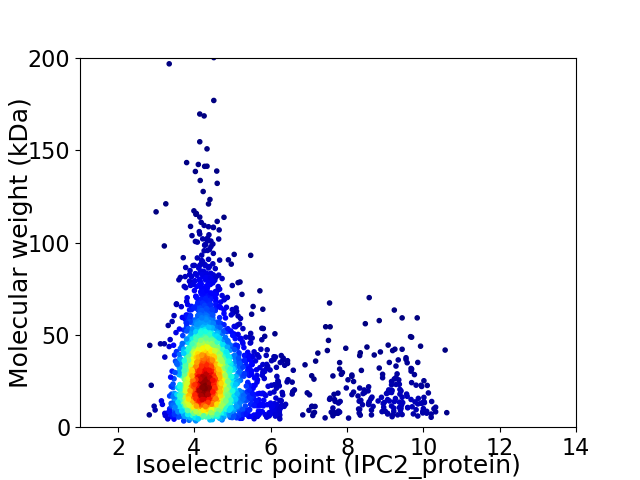
Virtual 2D-PAGE plot for 2931 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M0FG02|M0FG02_9EURY Nucleotide pyrophosphohydrolase OS=Halorubrum hochstenium ATCC 700873 OX=1227481 GN=C467_04515 PE=4 SV=1
MM1 pKa = 7.14VGAASAAPGNGGGPDD16 pKa = 3.05RR17 pKa = 11.84RR18 pKa = 11.84EE19 pKa = 4.07YY20 pKa = 10.56IVGVSATEE28 pKa = 4.44DD29 pKa = 3.5DD30 pKa = 3.71PKK32 pKa = 11.41GYY34 pKa = 10.5VKK36 pKa = 10.33QHH38 pKa = 5.71LKK40 pKa = 10.91GNEE43 pKa = 3.87EE44 pKa = 4.07VLSSNEE50 pKa = 4.05TLNTTVVGLPEE61 pKa = 4.14NAAEE65 pKa = 4.15PAHH68 pKa = 6.07EE69 pKa = 4.36NFRR72 pKa = 11.84EE73 pKa = 3.99RR74 pKa = 11.84QKK76 pKa = 10.97RR77 pKa = 11.84ADD79 pKa = 3.14AVKK82 pKa = 10.01YY83 pKa = 9.71VEE85 pKa = 4.18EE86 pKa = 4.19NVRR89 pKa = 11.84LTAQLAPNDD98 pKa = 4.09PQYY101 pKa = 11.64GDD103 pKa = 3.55QYY105 pKa = 11.89ADD107 pKa = 3.44QQVNAPDD114 pKa = 3.12AWEE117 pKa = 4.14EE118 pKa = 4.27TFGDD122 pKa = 3.83ANVTIGVVDD131 pKa = 3.41QGVKK135 pKa = 10.12YY136 pKa = 10.07DD137 pKa = 4.85HH138 pKa = 7.72PDD140 pKa = 3.1LDD142 pKa = 4.37GNMDD146 pKa = 4.26DD147 pKa = 5.15AVSNYY152 pKa = 9.7GRR154 pKa = 11.84DD155 pKa = 3.65FVDD158 pKa = 5.52DD159 pKa = 5.52DD160 pKa = 4.44GDD162 pKa = 4.18PYY164 pKa = 11.21PDD166 pKa = 4.0ALSDD170 pKa = 3.91EE171 pKa = 4.57YY172 pKa = 11.17HH173 pKa = 5.76GTHH176 pKa = 6.07VSGIAAAEE184 pKa = 4.02TDD186 pKa = 3.29NGEE189 pKa = 4.15GVTGIGNSSVLSGRR203 pKa = 11.84ALSEE207 pKa = 4.08RR208 pKa = 11.84GSGSTSDD215 pKa = 3.38IADD218 pKa = 3.64AVTWAADD225 pKa = 3.29QGADD229 pKa = 4.73VINLSLGGGGYY240 pKa = 8.89TDD242 pKa = 3.24TMKK245 pKa = 10.92NAVSYY250 pKa = 8.98ATDD253 pKa = 3.07KK254 pKa = 11.2GALVVAAAGNDD265 pKa = 3.14GRR267 pKa = 11.84NSVSYY272 pKa = 8.93PAAYY276 pKa = 9.87SEE278 pKa = 4.46CLGVSALDD286 pKa = 3.78PDD288 pKa = 3.99EE289 pKa = 4.65TLASYY294 pKa = 11.3SNYY297 pKa = 10.18GNEE300 pKa = 4.48IEE302 pKa = 4.78LAAPGTNVLSTWTDD316 pKa = 2.96DD317 pKa = 3.83GYY319 pKa = 11.68EE320 pKa = 4.4KK321 pKa = 10.5ISGTSMATPVVAGVAGLTLAQYY343 pKa = 11.29DD344 pKa = 3.92LTNAEE349 pKa = 4.63LRR351 pKa = 11.84DD352 pKa = 3.96HH353 pKa = 6.93LKK355 pKa = 9.49ATAVDD360 pKa = 3.68VGLSTEE366 pKa = 4.15KK367 pKa = 10.47QGSGRR372 pKa = 11.84VDD374 pKa = 2.95AGVAVTTDD382 pKa = 3.7PEE384 pKa = 4.47SGGGGGGGGGGSGGSTSTSVTGSLSDD410 pKa = 4.19YY411 pKa = 11.38SDD413 pKa = 4.05GDD415 pKa = 3.74CLRR418 pKa = 11.84YY419 pKa = 9.97AFEE422 pKa = 4.75YY423 pKa = 10.67GDD425 pKa = 3.73PSKK428 pKa = 11.35VVLDD432 pKa = 4.14LDD434 pKa = 4.42GPSDD438 pKa = 4.53ADD440 pKa = 3.18FDD442 pKa = 5.2LYY444 pKa = 11.65ANTGVDD450 pKa = 3.74ACPTTSDD457 pKa = 3.03YY458 pKa = 11.16DD459 pKa = 3.82YY460 pKa = 11.27RR461 pKa = 11.84SWTTDD466 pKa = 2.92SQEE469 pKa = 4.32TISIDD474 pKa = 3.72GPDD477 pKa = 3.52VSTDD481 pKa = 2.73LHH483 pKa = 7.7ALVDD487 pKa = 4.21SYY489 pKa = 11.64SGSGEE494 pKa = 3.89YY495 pKa = 10.15TLTITEE501 pKa = 4.32YY502 pKa = 10.63EE503 pKa = 4.08
MM1 pKa = 7.14VGAASAAPGNGGGPDD16 pKa = 3.05RR17 pKa = 11.84RR18 pKa = 11.84EE19 pKa = 4.07YY20 pKa = 10.56IVGVSATEE28 pKa = 4.44DD29 pKa = 3.5DD30 pKa = 3.71PKK32 pKa = 11.41GYY34 pKa = 10.5VKK36 pKa = 10.33QHH38 pKa = 5.71LKK40 pKa = 10.91GNEE43 pKa = 3.87EE44 pKa = 4.07VLSSNEE50 pKa = 4.05TLNTTVVGLPEE61 pKa = 4.14NAAEE65 pKa = 4.15PAHH68 pKa = 6.07EE69 pKa = 4.36NFRR72 pKa = 11.84EE73 pKa = 3.99RR74 pKa = 11.84QKK76 pKa = 10.97RR77 pKa = 11.84ADD79 pKa = 3.14AVKK82 pKa = 10.01YY83 pKa = 9.71VEE85 pKa = 4.18EE86 pKa = 4.19NVRR89 pKa = 11.84LTAQLAPNDD98 pKa = 4.09PQYY101 pKa = 11.64GDD103 pKa = 3.55QYY105 pKa = 11.89ADD107 pKa = 3.44QQVNAPDD114 pKa = 3.12AWEE117 pKa = 4.14EE118 pKa = 4.27TFGDD122 pKa = 3.83ANVTIGVVDD131 pKa = 3.41QGVKK135 pKa = 10.12YY136 pKa = 10.07DD137 pKa = 4.85HH138 pKa = 7.72PDD140 pKa = 3.1LDD142 pKa = 4.37GNMDD146 pKa = 4.26DD147 pKa = 5.15AVSNYY152 pKa = 9.7GRR154 pKa = 11.84DD155 pKa = 3.65FVDD158 pKa = 5.52DD159 pKa = 5.52DD160 pKa = 4.44GDD162 pKa = 4.18PYY164 pKa = 11.21PDD166 pKa = 4.0ALSDD170 pKa = 3.91EE171 pKa = 4.57YY172 pKa = 11.17HH173 pKa = 5.76GTHH176 pKa = 6.07VSGIAAAEE184 pKa = 4.02TDD186 pKa = 3.29NGEE189 pKa = 4.15GVTGIGNSSVLSGRR203 pKa = 11.84ALSEE207 pKa = 4.08RR208 pKa = 11.84GSGSTSDD215 pKa = 3.38IADD218 pKa = 3.64AVTWAADD225 pKa = 3.29QGADD229 pKa = 4.73VINLSLGGGGYY240 pKa = 8.89TDD242 pKa = 3.24TMKK245 pKa = 10.92NAVSYY250 pKa = 8.98ATDD253 pKa = 3.07KK254 pKa = 11.2GALVVAAAGNDD265 pKa = 3.14GRR267 pKa = 11.84NSVSYY272 pKa = 8.93PAAYY276 pKa = 9.87SEE278 pKa = 4.46CLGVSALDD286 pKa = 3.78PDD288 pKa = 3.99EE289 pKa = 4.65TLASYY294 pKa = 11.3SNYY297 pKa = 10.18GNEE300 pKa = 4.48IEE302 pKa = 4.78LAAPGTNVLSTWTDD316 pKa = 2.96DD317 pKa = 3.83GYY319 pKa = 11.68EE320 pKa = 4.4KK321 pKa = 10.5ISGTSMATPVVAGVAGLTLAQYY343 pKa = 11.29DD344 pKa = 3.92LTNAEE349 pKa = 4.63LRR351 pKa = 11.84DD352 pKa = 3.96HH353 pKa = 6.93LKK355 pKa = 9.49ATAVDD360 pKa = 3.68VGLSTEE366 pKa = 4.15KK367 pKa = 10.47QGSGRR372 pKa = 11.84VDD374 pKa = 2.95AGVAVTTDD382 pKa = 3.7PEE384 pKa = 4.47SGGGGGGGGGGSGGSTSTSVTGSLSDD410 pKa = 4.19YY411 pKa = 11.38SDD413 pKa = 4.05GDD415 pKa = 3.74CLRR418 pKa = 11.84YY419 pKa = 9.97AFEE422 pKa = 4.75YY423 pKa = 10.67GDD425 pKa = 3.73PSKK428 pKa = 11.35VVLDD432 pKa = 4.14LDD434 pKa = 4.42GPSDD438 pKa = 4.53ADD440 pKa = 3.18FDD442 pKa = 5.2LYY444 pKa = 11.65ANTGVDD450 pKa = 3.74ACPTTSDD457 pKa = 3.03YY458 pKa = 11.16DD459 pKa = 3.82YY460 pKa = 11.27RR461 pKa = 11.84SWTTDD466 pKa = 2.92SQEE469 pKa = 4.32TISIDD474 pKa = 3.72GPDD477 pKa = 3.52VSTDD481 pKa = 2.73LHH483 pKa = 7.7ALVDD487 pKa = 4.21SYY489 pKa = 11.64SGSGEE494 pKa = 3.89YY495 pKa = 10.15TLTITEE501 pKa = 4.32YY502 pKa = 10.63EE503 pKa = 4.08
Molecular weight: 52.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M0F001|M0F001_9EURY Glutamate--cysteine ligase OS=Halorubrum hochstenium ATCC 700873 OX=1227481 GN=gshA PE=3 SV=1
MM1 pKa = 7.49GMKK4 pKa = 10.47NNIGAADD11 pKa = 3.69RR12 pKa = 11.84RR13 pKa = 11.84IRR15 pKa = 11.84IGLGLGSAAVGLATLGGLLGLGTTVGAVLTLLGLVLVGTALVRR58 pKa = 11.84VCLLYY63 pKa = 10.81RR64 pKa = 11.84LLGVDD69 pKa = 3.62TSGSS73 pKa = 3.38
MM1 pKa = 7.49GMKK4 pKa = 10.47NNIGAADD11 pKa = 3.69RR12 pKa = 11.84RR13 pKa = 11.84IRR15 pKa = 11.84IGLGLGSAAVGLATLGGLLGLGTTVGAVLTLLGLVLVGTALVRR58 pKa = 11.84VCLLYY63 pKa = 10.81RR64 pKa = 11.84LLGVDD69 pKa = 3.62TSGSS73 pKa = 3.38
Molecular weight: 7.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
858121 |
31 |
1911 |
292.8 |
31.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.486 ± 0.077 | 0.635 ± 0.012 |
9.197 ± 0.062 | 8.491 ± 0.063 |
3.206 ± 0.031 | 9.281 ± 0.049 |
1.775 ± 0.023 | 3.457 ± 0.035 |
1.558 ± 0.025 | 8.531 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.565 ± 0.019 | 2.031 ± 0.023 |
4.867 ± 0.033 | 1.859 ± 0.024 |
6.988 ± 0.051 | 5.221 ± 0.037 |
5.938 ± 0.036 | 9.389 ± 0.045 |
1.046 ± 0.016 | 2.477 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
