
Molossus molossus circovirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 8.11
Get precalculated fractions of proteins
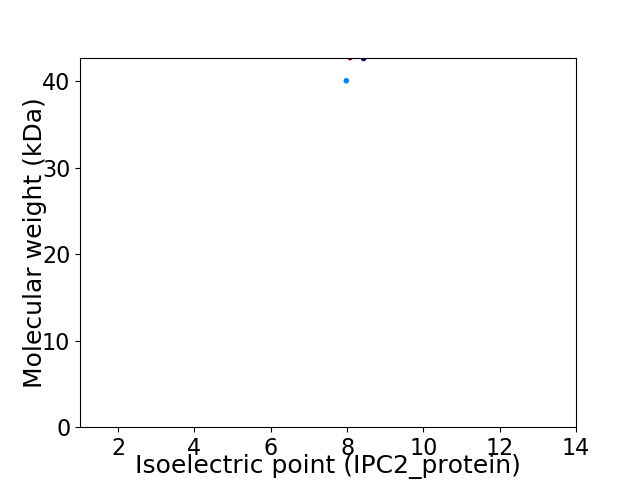
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I2MP64|A0A2I2MP64_9CIRC Capsid OS=Molossus molossus circovirus 2 OX=1959843 GN=Cap PE=3 SV=1
MM1 pKa = 7.39AAAPKK6 pKa = 9.93LVRR9 pKa = 11.84KK10 pKa = 9.82SRR12 pKa = 11.84FSNQEE17 pKa = 3.47NEE19 pKa = 4.0FDD21 pKa = 3.43RR22 pKa = 11.84GEE24 pKa = 4.04IGGSSRR30 pKa = 11.84SVCFTLNNYY39 pKa = 7.66TEE41 pKa = 4.34NDD43 pKa = 3.28LEE45 pKa = 4.67IIKK48 pKa = 8.64KK49 pKa = 6.5TWPKK53 pKa = 8.55IASYY57 pKa = 10.38YY58 pKa = 10.0VVGLEE63 pKa = 4.17VGEE66 pKa = 4.58SGTPHH71 pKa = 5.73LQGYY75 pKa = 7.28VEE77 pKa = 4.31WNSSKK82 pKa = 10.63EE83 pKa = 3.63FSTMHH88 pKa = 6.85KK89 pKa = 9.75IFSHH93 pKa = 5.79RR94 pKa = 11.84AHH96 pKa = 7.08WKK98 pKa = 9.56RR99 pKa = 11.84RR100 pKa = 11.84YY101 pKa = 9.35QNSTAEE107 pKa = 4.02QAAKK111 pKa = 9.92YY112 pKa = 8.9CKK114 pKa = 9.96KK115 pKa = 10.64GEE117 pKa = 3.97QSKK120 pKa = 10.64EE121 pKa = 3.63EE122 pKa = 4.09WEE124 pKa = 4.44KK125 pKa = 11.11FGSSGPNFGKK135 pKa = 9.92NANFFEE141 pKa = 5.4HH142 pKa = 7.35GEE144 pKa = 3.85ISNQGEE150 pKa = 3.98RR151 pKa = 11.84NDD153 pKa = 3.43IKK155 pKa = 10.94EE156 pKa = 4.23AMSMIKK162 pKa = 10.42NGATEE167 pKa = 3.86IEE169 pKa = 4.48VFEE172 pKa = 4.31ACPRR176 pKa = 11.84VSMQYY181 pKa = 10.82LKK183 pKa = 11.32GMDD186 pKa = 3.82RR187 pKa = 11.84YY188 pKa = 10.72RR189 pKa = 11.84LLCQKK194 pKa = 8.13QARR197 pKa = 11.84KK198 pKa = 10.12GYY200 pKa = 8.14NKK202 pKa = 10.44KK203 pKa = 10.04NVRR206 pKa = 11.84VYY208 pKa = 9.89WGEE211 pKa = 3.81TGTGKK216 pKa = 8.15TRR218 pKa = 11.84SALEE222 pKa = 4.0EE223 pKa = 4.15FPDD226 pKa = 5.14AFICTAGITGLWWDD240 pKa = 4.49GYY242 pKa = 10.97DD243 pKa = 3.96GEE245 pKa = 4.49EE246 pKa = 3.93TVIIDD251 pKa = 3.47EE252 pKa = 4.64FRR254 pKa = 11.84GNNCPLSMLLTILDD268 pKa = 4.87GYY270 pKa = 10.75ASQVSIRR277 pKa = 11.84GGSKK281 pKa = 9.28ILYY284 pKa = 9.16AKK286 pKa = 9.51TIIITSNIHH295 pKa = 5.38PDD297 pKa = 2.86EE298 pKa = 4.58WYY300 pKa = 10.99VNSDD304 pKa = 4.2DD305 pKa = 3.99YY306 pKa = 12.06SKK308 pKa = 11.33AALKK312 pKa = 10.51RR313 pKa = 11.84RR314 pKa = 11.84IDD316 pKa = 3.43STLFFSNKK324 pKa = 7.49EE325 pKa = 4.02NRR327 pKa = 11.84KK328 pKa = 8.29KK329 pKa = 8.83TAEE332 pKa = 3.98VEE334 pKa = 4.15GNTRR338 pKa = 11.84SSTSLEE344 pKa = 4.01KK345 pKa = 10.37FLKK348 pKa = 10.9LNLL351 pKa = 3.76
MM1 pKa = 7.39AAAPKK6 pKa = 9.93LVRR9 pKa = 11.84KK10 pKa = 9.82SRR12 pKa = 11.84FSNQEE17 pKa = 3.47NEE19 pKa = 4.0FDD21 pKa = 3.43RR22 pKa = 11.84GEE24 pKa = 4.04IGGSSRR30 pKa = 11.84SVCFTLNNYY39 pKa = 7.66TEE41 pKa = 4.34NDD43 pKa = 3.28LEE45 pKa = 4.67IIKK48 pKa = 8.64KK49 pKa = 6.5TWPKK53 pKa = 8.55IASYY57 pKa = 10.38YY58 pKa = 10.0VVGLEE63 pKa = 4.17VGEE66 pKa = 4.58SGTPHH71 pKa = 5.73LQGYY75 pKa = 7.28VEE77 pKa = 4.31WNSSKK82 pKa = 10.63EE83 pKa = 3.63FSTMHH88 pKa = 6.85KK89 pKa = 9.75IFSHH93 pKa = 5.79RR94 pKa = 11.84AHH96 pKa = 7.08WKK98 pKa = 9.56RR99 pKa = 11.84RR100 pKa = 11.84YY101 pKa = 9.35QNSTAEE107 pKa = 4.02QAAKK111 pKa = 9.92YY112 pKa = 8.9CKK114 pKa = 9.96KK115 pKa = 10.64GEE117 pKa = 3.97QSKK120 pKa = 10.64EE121 pKa = 3.63EE122 pKa = 4.09WEE124 pKa = 4.44KK125 pKa = 11.11FGSSGPNFGKK135 pKa = 9.92NANFFEE141 pKa = 5.4HH142 pKa = 7.35GEE144 pKa = 3.85ISNQGEE150 pKa = 3.98RR151 pKa = 11.84NDD153 pKa = 3.43IKK155 pKa = 10.94EE156 pKa = 4.23AMSMIKK162 pKa = 10.42NGATEE167 pKa = 3.86IEE169 pKa = 4.48VFEE172 pKa = 4.31ACPRR176 pKa = 11.84VSMQYY181 pKa = 10.82LKK183 pKa = 11.32GMDD186 pKa = 3.82RR187 pKa = 11.84YY188 pKa = 10.72RR189 pKa = 11.84LLCQKK194 pKa = 8.13QARR197 pKa = 11.84KK198 pKa = 10.12GYY200 pKa = 8.14NKK202 pKa = 10.44KK203 pKa = 10.04NVRR206 pKa = 11.84VYY208 pKa = 9.89WGEE211 pKa = 3.81TGTGKK216 pKa = 8.15TRR218 pKa = 11.84SALEE222 pKa = 4.0EE223 pKa = 4.15FPDD226 pKa = 5.14AFICTAGITGLWWDD240 pKa = 4.49GYY242 pKa = 10.97DD243 pKa = 3.96GEE245 pKa = 4.49EE246 pKa = 3.93TVIIDD251 pKa = 3.47EE252 pKa = 4.64FRR254 pKa = 11.84GNNCPLSMLLTILDD268 pKa = 4.87GYY270 pKa = 10.75ASQVSIRR277 pKa = 11.84GGSKK281 pKa = 9.28ILYY284 pKa = 9.16AKK286 pKa = 9.51TIIITSNIHH295 pKa = 5.38PDD297 pKa = 2.86EE298 pKa = 4.58WYY300 pKa = 10.99VNSDD304 pKa = 4.2DD305 pKa = 3.99YY306 pKa = 12.06SKK308 pKa = 11.33AALKK312 pKa = 10.51RR313 pKa = 11.84RR314 pKa = 11.84IDD316 pKa = 3.43STLFFSNKK324 pKa = 7.49EE325 pKa = 4.02NRR327 pKa = 11.84KK328 pKa = 8.29KK329 pKa = 8.83TAEE332 pKa = 3.98VEE334 pKa = 4.15GNTRR338 pKa = 11.84SSTSLEE344 pKa = 4.01KK345 pKa = 10.37FLKK348 pKa = 10.9LNLL351 pKa = 3.76
Molecular weight: 40.03 kDa
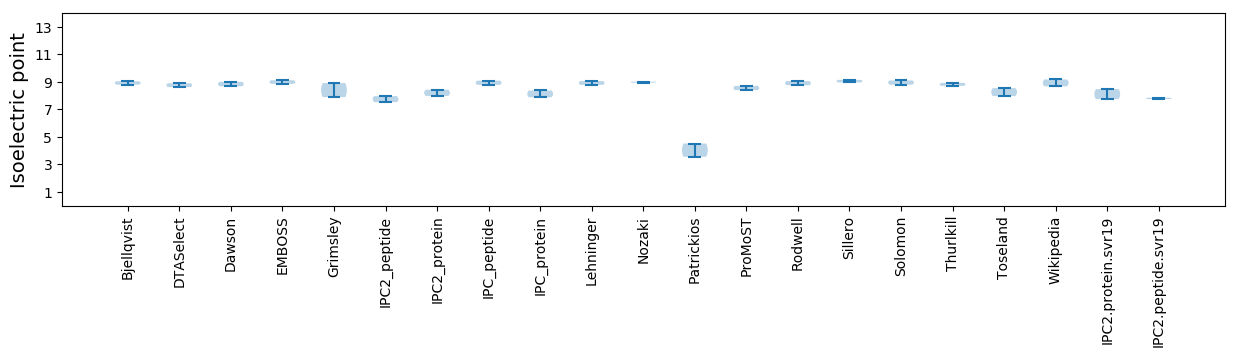
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I2MP64|A0A2I2MP64_9CIRC Capsid OS=Molossus molossus circovirus 2 OX=1959843 GN=Cap PE=3 SV=1
MM1 pKa = 7.42PAKK4 pKa = 10.37KK5 pKa = 9.68IYY7 pKa = 10.78NPAAYY12 pKa = 9.27HH13 pKa = 5.45AAVRR17 pKa = 11.84RR18 pKa = 11.84NAAAAKK24 pKa = 9.25SAAVAHH30 pKa = 5.97ARR32 pKa = 11.84KK33 pKa = 8.84HH34 pKa = 4.63ATHH37 pKa = 7.41FIQRR41 pKa = 11.84GVKK44 pKa = 9.66SADD47 pKa = 3.57RR48 pKa = 11.84AVKK51 pKa = 10.18KK52 pKa = 10.26YY53 pKa = 10.14VSSGAMSRR61 pKa = 11.84DD62 pKa = 3.4LQRR65 pKa = 11.84GARR68 pKa = 11.84AVTRR72 pKa = 11.84SAIMSGSGDD81 pKa = 3.56INIHH85 pKa = 5.44KK86 pKa = 10.58SKK88 pKa = 10.39LRR90 pKa = 11.84EE91 pKa = 3.68RR92 pKa = 11.84AYY94 pKa = 9.89DD95 pKa = 3.61TSGHH99 pKa = 5.13PTVRR103 pKa = 11.84LTEE106 pKa = 4.19TGEE109 pKa = 3.55MTIIHH114 pKa = 6.68RR115 pKa = 11.84EE116 pKa = 4.04YY117 pKa = 10.18IGEE120 pKa = 4.66LISGTQNPANFTSQNYY136 pKa = 9.56GINPGNPGCFPWLSTVSNSFQNYY159 pKa = 8.6KK160 pKa = 9.99FHH162 pKa = 7.11SLVFEE167 pKa = 4.14YY168 pKa = 11.18VPLTSEE174 pKa = 4.04STATSTGALVGMGSVMMATQYY195 pKa = 11.3DD196 pKa = 4.36SSLGPYY202 pKa = 9.61LNKK205 pKa = 10.08PQMEE209 pKa = 4.19NSDD212 pKa = 3.95FATSAKK218 pKa = 9.62PSSAMVHH225 pKa = 5.16VVEE228 pKa = 5.38CNSKK232 pKa = 9.59YY233 pKa = 10.69NVLGEE238 pKa = 4.5YY239 pKa = 9.99YY240 pKa = 10.4VSSDD244 pKa = 3.22TTVSNTNISNADD256 pKa = 2.7IRR258 pKa = 11.84MQNVGIFQIASVGIPTYY275 pKa = 10.87GSVAIDD281 pKa = 3.91LGEE284 pKa = 3.79IWVSYY289 pKa = 10.7SITLYY294 pKa = 10.83KK295 pKa = 8.59PTIYY299 pKa = 10.48GALAAIQSSHH309 pKa = 4.6YY310 pKa = 9.63TNGAVSGAPTSANPFGTVTTSNQPTPATNNQLPLTFTSGSTFAFPLAITEE360 pKa = 4.1GNYY363 pKa = 10.54LIVYY367 pKa = 8.78QCVSGNAVTFSFTGIVTTNCTALTCWSTPMTT398 pKa = 4.35
MM1 pKa = 7.42PAKK4 pKa = 10.37KK5 pKa = 9.68IYY7 pKa = 10.78NPAAYY12 pKa = 9.27HH13 pKa = 5.45AAVRR17 pKa = 11.84RR18 pKa = 11.84NAAAAKK24 pKa = 9.25SAAVAHH30 pKa = 5.97ARR32 pKa = 11.84KK33 pKa = 8.84HH34 pKa = 4.63ATHH37 pKa = 7.41FIQRR41 pKa = 11.84GVKK44 pKa = 9.66SADD47 pKa = 3.57RR48 pKa = 11.84AVKK51 pKa = 10.18KK52 pKa = 10.26YY53 pKa = 10.14VSSGAMSRR61 pKa = 11.84DD62 pKa = 3.4LQRR65 pKa = 11.84GARR68 pKa = 11.84AVTRR72 pKa = 11.84SAIMSGSGDD81 pKa = 3.56INIHH85 pKa = 5.44KK86 pKa = 10.58SKK88 pKa = 10.39LRR90 pKa = 11.84EE91 pKa = 3.68RR92 pKa = 11.84AYY94 pKa = 9.89DD95 pKa = 3.61TSGHH99 pKa = 5.13PTVRR103 pKa = 11.84LTEE106 pKa = 4.19TGEE109 pKa = 3.55MTIIHH114 pKa = 6.68RR115 pKa = 11.84EE116 pKa = 4.04YY117 pKa = 10.18IGEE120 pKa = 4.66LISGTQNPANFTSQNYY136 pKa = 9.56GINPGNPGCFPWLSTVSNSFQNYY159 pKa = 8.6KK160 pKa = 9.99FHH162 pKa = 7.11SLVFEE167 pKa = 4.14YY168 pKa = 11.18VPLTSEE174 pKa = 4.04STATSTGALVGMGSVMMATQYY195 pKa = 11.3DD196 pKa = 4.36SSLGPYY202 pKa = 9.61LNKK205 pKa = 10.08PQMEE209 pKa = 4.19NSDD212 pKa = 3.95FATSAKK218 pKa = 9.62PSSAMVHH225 pKa = 5.16VVEE228 pKa = 5.38CNSKK232 pKa = 9.59YY233 pKa = 10.69NVLGEE238 pKa = 4.5YY239 pKa = 9.99YY240 pKa = 10.4VSSDD244 pKa = 3.22TTVSNTNISNADD256 pKa = 2.7IRR258 pKa = 11.84MQNVGIFQIASVGIPTYY275 pKa = 10.87GSVAIDD281 pKa = 3.91LGEE284 pKa = 3.79IWVSYY289 pKa = 10.7SITLYY294 pKa = 10.83KK295 pKa = 8.59PTIYY299 pKa = 10.48GALAAIQSSHH309 pKa = 4.6YY310 pKa = 9.63TNGAVSGAPTSANPFGTVTTSNQPTPATNNQLPLTFTSGSTFAFPLAITEE360 pKa = 4.1GNYY363 pKa = 10.54LIVYY367 pKa = 8.78QCVSGNAVTFSFTGIVTTNCTALTCWSTPMTT398 pKa = 4.35
Molecular weight: 42.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
749 |
351 |
398 |
374.5 |
41.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.411 ± 1.647 | 1.469 ± 0.163 |
2.937 ± 0.52 | 5.874 ± 2.199 |
4.005 ± 0.375 | 7.61 ± 0.249 |
2.136 ± 0.289 | 5.874 ± 0.074 |
6.008 ± 1.915 | 5.34 ± 0.436 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.403 ± 0.277 | 6.542 ± 0.007 |
3.738 ± 0.989 | 3.071 ± 0.15 |
4.539 ± 0.786 | 10.147 ± 0.892 |
8.144 ± 1.659 | 5.607 ± 1.098 |
1.469 ± 0.55 | 4.673 ± 0.271 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
