
Tortoise microvirus 68
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
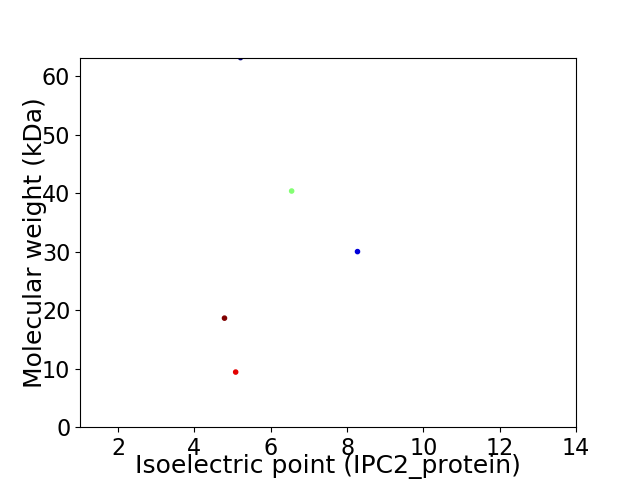
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FW03|A0A4V1FW03_9VIRU Internal scaffolding protein OS=Tortoise microvirus 68 OX=2583174 PE=4 SV=1
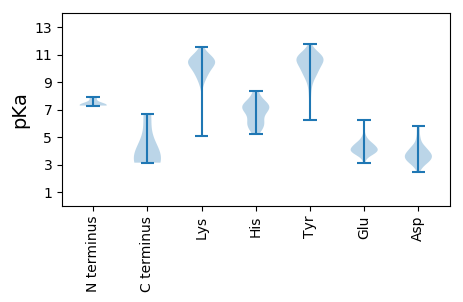
MM1 pKa = 7.31EE2 pKa = 6.24AISFEE7 pKa = 4.15TQYY10 pKa = 11.1RR11 pKa = 11.84KK12 pKa = 9.72PKK14 pKa = 10.48RR15 pKa = 11.84FFTSEE20 pKa = 3.79GSPIKK25 pKa = 9.45ITYY28 pKa = 9.58GAVFDD33 pKa = 4.1NKK35 pKa = 10.37GRR37 pKa = 11.84LQLEE41 pKa = 4.51EE42 pKa = 4.56KK43 pKa = 10.67GSEE46 pKa = 3.91NLYY49 pKa = 10.93DD50 pKa = 4.07YY51 pKa = 10.21IQSFAEE57 pKa = 4.03SVDD60 pKa = 2.76IHH62 pKa = 7.1VILKK66 pKa = 10.26KK67 pKa = 10.22FANGDD72 pKa = 3.62STVLSRR78 pKa = 11.84AQGFYY83 pKa = 11.22GDD85 pKa = 3.69VTEE88 pKa = 6.12FPTTYY93 pKa = 11.01ADD95 pKa = 3.23VLNRR99 pKa = 11.84VIEE102 pKa = 4.26GEE104 pKa = 4.11NFFNTLPVEE113 pKa = 4.31TRR115 pKa = 11.84SKK117 pKa = 10.64FNHH120 pKa = 6.44SYY122 pKa = 11.14SEE124 pKa = 4.65FIASIGSQQFFDD136 pKa = 3.97AFGMSKK142 pKa = 10.47PSDD145 pKa = 3.68QNVVDD150 pKa = 5.06PVDD153 pKa = 4.09LPDD156 pKa = 3.36VVKK159 pKa = 10.39EE160 pKa = 4.26VKK162 pKa = 10.44SDD164 pKa = 3.43VV165 pKa = 3.41
MM1 pKa = 7.31EE2 pKa = 6.24AISFEE7 pKa = 4.15TQYY10 pKa = 11.1RR11 pKa = 11.84KK12 pKa = 9.72PKK14 pKa = 10.48RR15 pKa = 11.84FFTSEE20 pKa = 3.79GSPIKK25 pKa = 9.45ITYY28 pKa = 9.58GAVFDD33 pKa = 4.1NKK35 pKa = 10.37GRR37 pKa = 11.84LQLEE41 pKa = 4.51EE42 pKa = 4.56KK43 pKa = 10.67GSEE46 pKa = 3.91NLYY49 pKa = 10.93DD50 pKa = 4.07YY51 pKa = 10.21IQSFAEE57 pKa = 4.03SVDD60 pKa = 2.76IHH62 pKa = 7.1VILKK66 pKa = 10.26KK67 pKa = 10.22FANGDD72 pKa = 3.62STVLSRR78 pKa = 11.84AQGFYY83 pKa = 11.22GDD85 pKa = 3.69VTEE88 pKa = 6.12FPTTYY93 pKa = 11.01ADD95 pKa = 3.23VLNRR99 pKa = 11.84VIEE102 pKa = 4.26GEE104 pKa = 4.11NFFNTLPVEE113 pKa = 4.31TRR115 pKa = 11.84SKK117 pKa = 10.64FNHH120 pKa = 6.44SYY122 pKa = 11.14SEE124 pKa = 4.65FIASIGSQQFFDD136 pKa = 3.97AFGMSKK142 pKa = 10.47PSDD145 pKa = 3.68QNVVDD150 pKa = 5.06PVDD153 pKa = 4.09LPDD156 pKa = 3.36VVKK159 pKa = 10.39EE160 pKa = 4.26VKK162 pKa = 10.44SDD164 pKa = 3.43VV165 pKa = 3.41
Molecular weight: 18.65 kDa
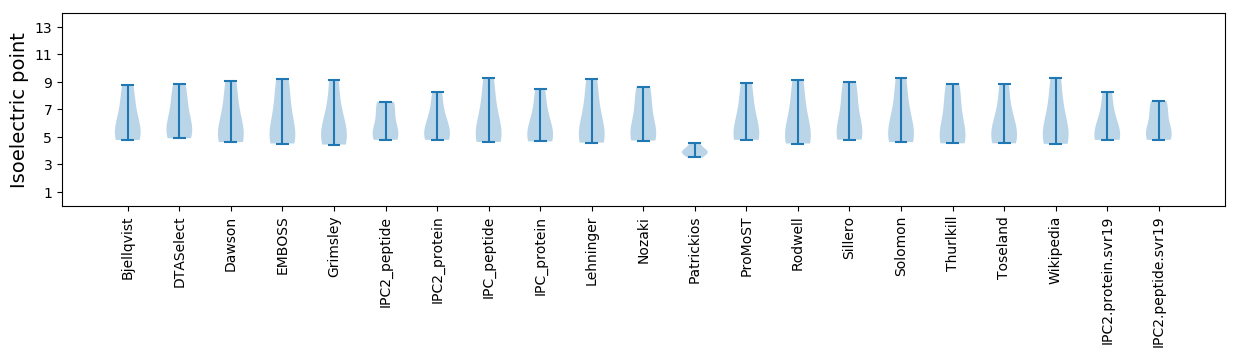
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7D7|A0A4P8W7D7_9VIRU Replication initiation protein OS=Tortoise microvirus 68 OX=2583174 PE=4 SV=1
MM1 pKa = 7.29SVSNGISTYY10 pKa = 10.3TPAPANQNMIQAVNSAANNIGMMQGIAQANSAASARR46 pKa = 11.84EE47 pKa = 3.67ARR49 pKa = 11.84IQRR52 pKa = 11.84EE53 pKa = 3.7WQEE56 pKa = 4.09RR57 pKa = 11.84QNARR61 pKa = 11.84AMAFNSLEE69 pKa = 4.0ASKK72 pKa = 11.11SRR74 pKa = 11.84DD75 pKa = 3.12WQQMMSNTAHH85 pKa = 5.68QRR87 pKa = 11.84EE88 pKa = 4.5IQDD91 pKa = 3.91LMAAGLNPVLSAMGGNGAAVTSGATASGVTSAGAKK126 pKa = 10.13GDD128 pKa = 3.31VDD130 pKa = 3.97TSANAAITNLLGSLLSAEE148 pKa = 4.46TSIKK152 pKa = 10.04AANINAQTQSAVADD166 pKa = 4.24KK167 pKa = 8.93YY168 pKa = 9.39TAMSQLVAEE177 pKa = 4.94LNAAASMYY185 pKa = 10.71SADD188 pKa = 3.44SHH190 pKa = 7.29AGASRR195 pKa = 11.84YY196 pKa = 9.68AADD199 pKa = 3.86RR200 pKa = 11.84SAAASMFGSSLMSAASKK217 pKa = 10.26HH218 pKa = 6.09AADD221 pKa = 3.73QSYY224 pKa = 9.42EE225 pKa = 3.73ASRR228 pKa = 11.84YY229 pKa = 7.65RR230 pKa = 11.84ANMDD234 pKa = 2.96WSITNEE240 pKa = 4.03KK241 pKa = 9.63NIHH244 pKa = 5.69DD245 pKa = 5.54AIVKK249 pKa = 9.95DD250 pKa = 3.96LYY252 pKa = 11.6GNNQYY257 pKa = 9.95TGIFGWILSKK267 pKa = 10.58WPIFDD272 pKa = 3.9YY273 pKa = 10.74SSQRR277 pKa = 11.84RR278 pKa = 11.84MFSKK282 pKa = 10.7RR283 pKa = 3.14
MM1 pKa = 7.29SVSNGISTYY10 pKa = 10.3TPAPANQNMIQAVNSAANNIGMMQGIAQANSAASARR46 pKa = 11.84EE47 pKa = 3.67ARR49 pKa = 11.84IQRR52 pKa = 11.84EE53 pKa = 3.7WQEE56 pKa = 4.09RR57 pKa = 11.84QNARR61 pKa = 11.84AMAFNSLEE69 pKa = 4.0ASKK72 pKa = 11.11SRR74 pKa = 11.84DD75 pKa = 3.12WQQMMSNTAHH85 pKa = 5.68QRR87 pKa = 11.84EE88 pKa = 4.5IQDD91 pKa = 3.91LMAAGLNPVLSAMGGNGAAVTSGATASGVTSAGAKK126 pKa = 10.13GDD128 pKa = 3.31VDD130 pKa = 3.97TSANAAITNLLGSLLSAEE148 pKa = 4.46TSIKK152 pKa = 10.04AANINAQTQSAVADD166 pKa = 4.24KK167 pKa = 8.93YY168 pKa = 9.39TAMSQLVAEE177 pKa = 4.94LNAAASMYY185 pKa = 10.71SADD188 pKa = 3.44SHH190 pKa = 7.29AGASRR195 pKa = 11.84YY196 pKa = 9.68AADD199 pKa = 3.86RR200 pKa = 11.84SAAASMFGSSLMSAASKK217 pKa = 10.26HH218 pKa = 6.09AADD221 pKa = 3.73QSYY224 pKa = 9.42EE225 pKa = 3.73ASRR228 pKa = 11.84YY229 pKa = 7.65RR230 pKa = 11.84ANMDD234 pKa = 2.96WSITNEE240 pKa = 4.03KK241 pKa = 9.63NIHH244 pKa = 5.69DD245 pKa = 5.54AIVKK249 pKa = 9.95DD250 pKa = 3.96LYY252 pKa = 11.6GNNQYY257 pKa = 9.95TGIFGWILSKK267 pKa = 10.58WPIFDD272 pKa = 3.9YY273 pKa = 10.74SSQRR277 pKa = 11.84RR278 pKa = 11.84MFSKK282 pKa = 10.7RR283 pKa = 3.14
Molecular weight: 30.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1446 |
83 |
568 |
289.2 |
32.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.437 ± 2.204 | 0.899 ± 0.559 |
6.432 ± 0.543 | 4.91 ± 1.039 |
4.703 ± 0.833 | 6.155 ± 0.176 |
1.936 ± 0.263 | 5.533 ± 0.46 |
4.495 ± 1.003 | 6.777 ± 0.656 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.702 | 4.979 ± 0.607 |
4.011 ± 0.769 | 4.219 ± 0.733 |
5.394 ± 0.542 | 9.959 ± 0.979 |
5.878 ± 0.912 | 6.501 ± 1.465 |
1.383 ± 0.399 | 4.979 ± 0.591 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
