
Ketogulonicigenium vulgare (strain WSH-001)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Ketogulonicigenium; Ketogulonicigenium vulgare
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
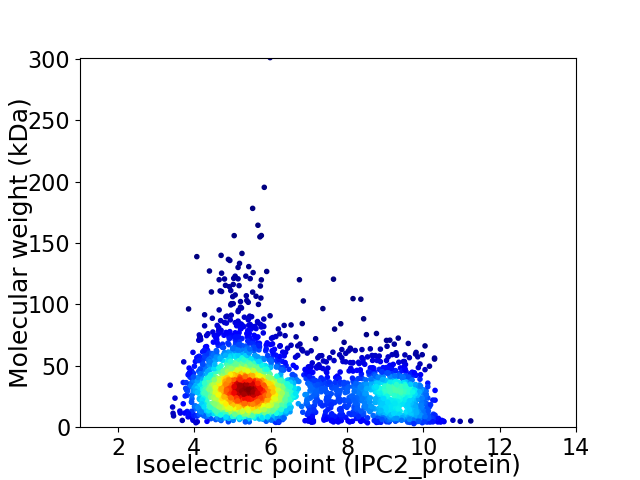
Virtual 2D-PAGE plot for 3049 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F9Y6Q7|F9Y6Q7_KETVW Isochorismatase hydrolase OS=Ketogulonicigenium vulgare (strain WSH-001) OX=759362 GN=pncA PE=4 SV=1
MM1 pKa = 7.48LGKK4 pKa = 10.19LAFSGMQHH12 pKa = 6.41EE13 pKa = 4.73GHH15 pKa = 5.65FMKK18 pKa = 10.89RR19 pKa = 11.84ILLASAALTAFAGAAAAEE37 pKa = 4.21ISFNGDD43 pKa = 2.29ARR45 pKa = 11.84LGYY48 pKa = 10.58NDD50 pKa = 3.84TAIEE54 pKa = 3.83NDD56 pKa = 3.08GFYY59 pKa = 10.34WRR61 pKa = 11.84LRR63 pKa = 11.84MNIAASQEE71 pKa = 4.0LDD73 pKa = 3.06NGLTFGGSVRR83 pKa = 11.84FDD85 pKa = 4.97LIDD88 pKa = 3.44NGYY91 pKa = 10.09ISDD94 pKa = 4.48PALDD98 pKa = 3.89TNNLKK103 pKa = 10.77LFLEE107 pKa = 4.5AGEE110 pKa = 4.42LGGLYY115 pKa = 10.5FGDD118 pKa = 4.17YY119 pKa = 10.32EE120 pKa = 4.69FSAVDD125 pKa = 3.08AWKK128 pKa = 10.22AAGPMFADD136 pKa = 5.09GFSEE140 pKa = 4.19QDD142 pKa = 3.09GEE144 pKa = 4.24KK145 pKa = 9.81TLRR148 pKa = 11.84GDD150 pKa = 3.16INYY153 pKa = 10.01GNVAVSVSTPISSTTGNDD171 pKa = 3.21NASEE175 pKa = 4.21SQLGQWNFAASTTFGAFDD193 pKa = 3.8VVFAYY198 pKa = 10.23QGEE201 pKa = 4.23GDD203 pKa = 3.88DD204 pKa = 6.02DD205 pKa = 4.58YY206 pKa = 11.55FAEE209 pKa = 4.76AALGDD214 pKa = 4.74DD215 pKa = 4.06INPNEE220 pKa = 4.49IIGLSVGTTFAGASVRR236 pKa = 11.84LAYY239 pKa = 8.67ATTDD243 pKa = 3.39YY244 pKa = 10.93KK245 pKa = 11.38VAGTTQSSLGLTATYY260 pKa = 10.34PLGPVALTGWVIFEE274 pKa = 5.4DD275 pKa = 4.64GDD277 pKa = 3.49VSDD280 pKa = 6.17AMDD283 pKa = 3.97EE284 pKa = 4.09DD285 pKa = 4.47LFYY288 pKa = 11.01GARR291 pKa = 11.84VAYY294 pKa = 9.31AANGFSISADD304 pKa = 3.4YY305 pKa = 11.29GEE307 pKa = 4.93RR308 pKa = 11.84LANNLSIGGRR318 pKa = 11.84DD319 pKa = 3.13ARR321 pKa = 11.84LAVEE325 pKa = 4.69GGYY328 pKa = 10.83DD329 pKa = 3.62LGNGINLKK337 pKa = 10.4AGYY340 pKa = 9.73LSRR343 pKa = 11.84DD344 pKa = 2.94SWSIGSGEE352 pKa = 4.06AYY354 pKa = 10.44YY355 pKa = 11.12VGGTYY360 pKa = 10.78DD361 pKa = 3.98LGAGATLLVSYY372 pKa = 10.99ADD374 pKa = 4.59ADD376 pKa = 4.01DD377 pKa = 4.0TSAGYY382 pKa = 10.78LSDD385 pKa = 5.18DD386 pKa = 3.8EE387 pKa = 4.93VGAPAYY393 pKa = 10.45QNGATIEE400 pKa = 4.12VSFKK404 pKa = 10.8FF405 pKa = 3.84
MM1 pKa = 7.48LGKK4 pKa = 10.19LAFSGMQHH12 pKa = 6.41EE13 pKa = 4.73GHH15 pKa = 5.65FMKK18 pKa = 10.89RR19 pKa = 11.84ILLASAALTAFAGAAAAEE37 pKa = 4.21ISFNGDD43 pKa = 2.29ARR45 pKa = 11.84LGYY48 pKa = 10.58NDD50 pKa = 3.84TAIEE54 pKa = 3.83NDD56 pKa = 3.08GFYY59 pKa = 10.34WRR61 pKa = 11.84LRR63 pKa = 11.84MNIAASQEE71 pKa = 4.0LDD73 pKa = 3.06NGLTFGGSVRR83 pKa = 11.84FDD85 pKa = 4.97LIDD88 pKa = 3.44NGYY91 pKa = 10.09ISDD94 pKa = 4.48PALDD98 pKa = 3.89TNNLKK103 pKa = 10.77LFLEE107 pKa = 4.5AGEE110 pKa = 4.42LGGLYY115 pKa = 10.5FGDD118 pKa = 4.17YY119 pKa = 10.32EE120 pKa = 4.69FSAVDD125 pKa = 3.08AWKK128 pKa = 10.22AAGPMFADD136 pKa = 5.09GFSEE140 pKa = 4.19QDD142 pKa = 3.09GEE144 pKa = 4.24KK145 pKa = 9.81TLRR148 pKa = 11.84GDD150 pKa = 3.16INYY153 pKa = 10.01GNVAVSVSTPISSTTGNDD171 pKa = 3.21NASEE175 pKa = 4.21SQLGQWNFAASTTFGAFDD193 pKa = 3.8VVFAYY198 pKa = 10.23QGEE201 pKa = 4.23GDD203 pKa = 3.88DD204 pKa = 6.02DD205 pKa = 4.58YY206 pKa = 11.55FAEE209 pKa = 4.76AALGDD214 pKa = 4.74DD215 pKa = 4.06INPNEE220 pKa = 4.49IIGLSVGTTFAGASVRR236 pKa = 11.84LAYY239 pKa = 8.67ATTDD243 pKa = 3.39YY244 pKa = 10.93KK245 pKa = 11.38VAGTTQSSLGLTATYY260 pKa = 10.34PLGPVALTGWVIFEE274 pKa = 5.4DD275 pKa = 4.64GDD277 pKa = 3.49VSDD280 pKa = 6.17AMDD283 pKa = 3.97EE284 pKa = 4.09DD285 pKa = 4.47LFYY288 pKa = 11.01GARR291 pKa = 11.84VAYY294 pKa = 9.31AANGFSISADD304 pKa = 3.4YY305 pKa = 11.29GEE307 pKa = 4.93RR308 pKa = 11.84LANNLSIGGRR318 pKa = 11.84DD319 pKa = 3.13ARR321 pKa = 11.84LAVEE325 pKa = 4.69GGYY328 pKa = 10.83DD329 pKa = 3.62LGNGINLKK337 pKa = 10.4AGYY340 pKa = 9.73LSRR343 pKa = 11.84DD344 pKa = 2.94SWSIGSGEE352 pKa = 4.06AYY354 pKa = 10.44YY355 pKa = 11.12VGGTYY360 pKa = 10.78DD361 pKa = 3.98LGAGATLLVSYY372 pKa = 10.99ADD374 pKa = 4.59ADD376 pKa = 4.01DD377 pKa = 4.0TSAGYY382 pKa = 10.78LSDD385 pKa = 5.18DD386 pKa = 3.8EE387 pKa = 4.93VGAPAYY393 pKa = 10.45QNGATIEE400 pKa = 4.12VSFKK404 pKa = 10.8FF405 pKa = 3.84
Molecular weight: 42.62 kDa
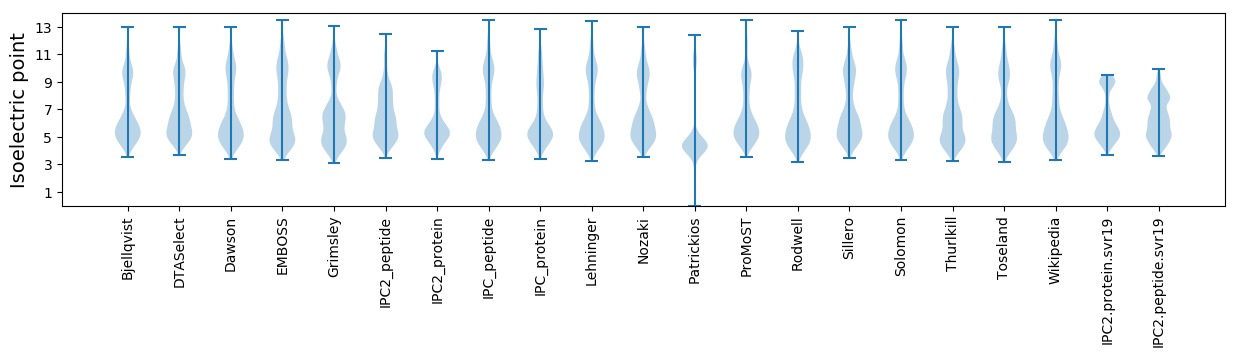
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F9Y5F4|F9Y5F4_KETVW Periplasmic protein thiol:disulfide oxidoreductase DsbE subfamily protein OS=Ketogulonicigenium vulgare (strain WSH-001) OX=759362 GN=cycY PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 9.2ARR15 pKa = 11.84HH16 pKa = 4.71GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97VLSAA44 pKa = 4.11
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 9.2ARR15 pKa = 11.84HH16 pKa = 4.71GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97VLSAA44 pKa = 4.11
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
950172 |
23 |
2789 |
311.6 |
33.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.121 ± 0.074 | 0.71 ± 0.014 |
5.962 ± 0.036 | 4.955 ± 0.038 |
3.682 ± 0.027 | 8.591 ± 0.04 |
2.065 ± 0.022 | 5.554 ± 0.029 |
2.514 ± 0.035 | 10.347 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.798 ± 0.022 | 2.659 ± 0.029 |
5.231 ± 0.031 | 3.615 ± 0.022 |
6.798 ± 0.042 | 5.066 ± 0.033 |
5.553 ± 0.031 | 7.183 ± 0.034 |
1.354 ± 0.019 | 2.242 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
