
Xanthobacter tagetidis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Xanthobacteraceae; Xanthobacter
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
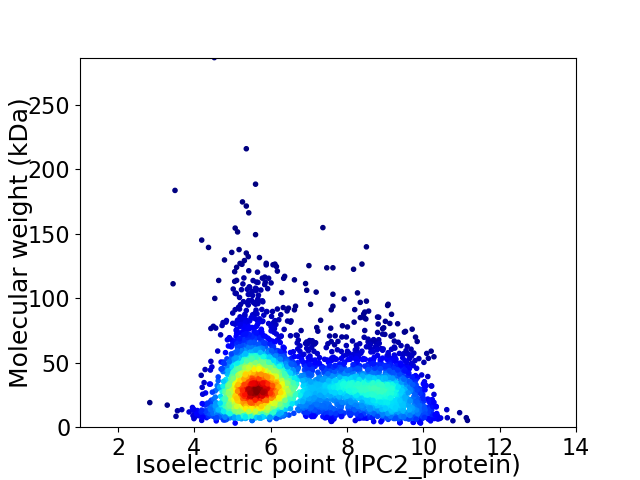
Virtual 2D-PAGE plot for 4442 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3L7A1J7|A0A3L7A1J7_9RHIZ Urease subunit gamma OS=Xanthobacter tagetidis OX=60216 GN=D9R14_19810 PE=3 SV=1
TT1 pKa = 6.13TAEE4 pKa = 4.26DD5 pKa = 4.01TPLVIAAADD14 pKa = 4.02LLANDD19 pKa = 4.79ADD21 pKa = 4.04IDD23 pKa = 4.24GDD25 pKa = 4.12TLSVTGVGGAVGGTVALDD43 pKa = 3.73DD44 pKa = 4.5GEE46 pKa = 4.52ITFTPDD52 pKa = 3.01ADD54 pKa = 4.24FNGDD58 pKa = 3.07ASFTYY63 pKa = 9.55TVSDD67 pKa = 3.78GKK69 pKa = 11.16GGTDD73 pKa = 2.94TATVNVTVTPVNDD86 pKa = 3.85APVAAADD93 pKa = 3.79TATTAEE99 pKa = 4.31DD100 pKa = 4.04TPLVIAAADD109 pKa = 4.02LLANDD114 pKa = 4.79ADD116 pKa = 4.04IDD118 pKa = 4.24GDD120 pKa = 4.12TLSVTGVGGAVGGTVALDD138 pKa = 3.83DD139 pKa = 4.66GEE141 pKa = 4.76VTFTPDD147 pKa = 3.42ANFNGDD153 pKa = 3.04ASFTYY158 pKa = 9.55TVSDD162 pKa = 3.78GKK164 pKa = 11.16GGTDD168 pKa = 2.94TATVNVTVTPVNDD181 pKa = 3.85APVAAADD188 pKa = 3.79TATTAEE194 pKa = 4.31DD195 pKa = 4.04TPLVIAAADD204 pKa = 4.02LLANDD209 pKa = 4.79ADD211 pKa = 4.04IDD213 pKa = 4.24GDD215 pKa = 4.12TLSVTGVGGAVGGTVALDD233 pKa = 3.83DD234 pKa = 4.66GEE236 pKa = 4.76VTFTPDD242 pKa = 3.42ANFNGDD248 pKa = 3.04ASFTYY253 pKa = 9.55TVSDD257 pKa = 3.78GKK259 pKa = 11.16GGTDD263 pKa = 2.94TATVNVTVTPVNDD276 pKa = 3.74APIATADD283 pKa = 3.57TATTAEE289 pKa = 4.31DD290 pKa = 4.14TPLVIAVADD299 pKa = 4.29LLANDD304 pKa = 4.6SDD306 pKa = 5.16LDD308 pKa = 4.06GDD310 pKa = 4.22TLSVTGVGGAVGGTVALDD328 pKa = 3.73DD329 pKa = 4.5GEE331 pKa = 4.52ITFTPDD337 pKa = 3.01ADD339 pKa = 4.24FNGDD343 pKa = 3.01ASFSYY348 pKa = 9.9TVSDD352 pKa = 3.88GKK354 pKa = 11.18GGTDD358 pKa = 3.11TATVGVTVTPVNDD371 pKa = 3.73APVVASGATATFAEE385 pKa = 4.62NGTGAVYY392 pKa = 10.33QIAASDD398 pKa = 4.07AEE400 pKa = 4.67DD401 pKa = 5.25DD402 pKa = 4.09GLTYY406 pKa = 11.06ALSGADD412 pKa = 3.29ADD414 pKa = 4.86LFTVDD419 pKa = 4.79ADD421 pKa = 3.69GKK423 pKa = 9.95VAFKK427 pKa = 10.52AAPDD431 pKa = 3.63VEE433 pKa = 4.77APADD437 pKa = 3.64AGGDD441 pKa = 3.45NVYY444 pKa = 11.09DD445 pKa = 3.74LVVEE449 pKa = 4.43VSDD452 pKa = 4.56GTDD455 pKa = 3.16TTSKK459 pKa = 10.78AVAITVTDD467 pKa = 3.61VAEE470 pKa = 4.31NSAPRR475 pKa = 11.84LVSAPVLGGITAEE488 pKa = 4.02AAAVGNGFSSSARR501 pKa = 11.84FSPNGTKK508 pKa = 10.34VLFQSTASNLVEE520 pKa = 4.24GDD522 pKa = 3.52ANGTEE527 pKa = 4.31DD528 pKa = 4.81VFVKK532 pKa = 10.6DD533 pKa = 4.38LEE535 pKa = 4.63TGAVTLVSSSSSGTPGNGWSNGHH558 pKa = 5.75QFSADD563 pKa = 3.1GTKK566 pKa = 10.55VLFYY570 pKa = 10.68SYY572 pKa = 11.27ASNLVAGDD580 pKa = 3.5INGAADD586 pKa = 3.86VFVKK590 pKa = 10.61DD591 pKa = 4.39LEE593 pKa = 4.63TGAVTLVSSSSSGTPGNGEE612 pKa = 3.66SSGYY616 pKa = 9.64QFSADD621 pKa = 3.2GTKK624 pKa = 10.55VLFYY628 pKa = 10.83SYY630 pKa = 11.43ANNLVAGDD638 pKa = 3.84INGVADD644 pKa = 3.75VFVKK648 pKa = 10.69NLEE651 pKa = 4.25TGAVTLVSSSSSGTPGNGTSSGYY674 pKa = 9.85QFSADD679 pKa = 3.2GTKK682 pKa = 10.55VLFYY686 pKa = 10.83SYY688 pKa = 11.43ANNLVAGDD696 pKa = 3.84INGVADD702 pKa = 3.75VFVKK706 pKa = 10.69NLEE709 pKa = 4.25TGAVTLVSSSSSGTPGNGTSYY730 pKa = 10.69GHH732 pKa = 6.03QFSADD737 pKa = 3.18GTKK740 pKa = 10.55VLFYY744 pKa = 10.83SYY746 pKa = 11.43ANNLVAGDD754 pKa = 3.84INGVADD760 pKa = 3.75VFVKK764 pKa = 10.69NLEE767 pKa = 4.25TGAVTLVSSSSSGTPGNGTSYY788 pKa = 10.7GYY790 pKa = 10.37QYY792 pKa = 11.09SADD795 pKa = 3.58GTKK798 pKa = 10.58VLFQSDD804 pKa = 3.69ASNLVAGDD812 pKa = 3.5INGAADD818 pKa = 3.69IFVKK822 pKa = 10.61DD823 pKa = 4.06LEE825 pKa = 4.72TGVVTLVSSSSSGTPGNQTSYY846 pKa = 10.27SHH848 pKa = 6.19QFSADD853 pKa = 3.09GTKK856 pKa = 10.47VLFYY860 pKa = 10.89SSASNLVAGDD870 pKa = 3.5INGAADD876 pKa = 3.86VFVKK880 pKa = 10.53DD881 pKa = 4.33LEE883 pKa = 4.74SGAVTLVSSSSSGTPGNGTSYY904 pKa = 10.69GHH906 pKa = 6.21QFSADD911 pKa = 3.35CAKK914 pKa = 10.82VLFRR918 pKa = 11.84SYY920 pKa = 11.39ASNLVAGDD928 pKa = 3.6TNGTYY933 pKa = 10.75DD934 pKa = 4.52VFMKK938 pKa = 10.72DD939 pKa = 3.12LATGEE944 pKa = 4.33VTLLSARR951 pKa = 11.84SLAATPLAKK960 pKa = 10.31SGAFTFDD967 pKa = 3.91DD968 pKa = 4.07ADD970 pKa = 4.01GGDD973 pKa = 3.38THH975 pKa = 6.5TASVLPAAGTLGTLQADD992 pKa = 3.52IVVVADD998 pKa = 3.94GPDD1001 pKa = 3.29RR1002 pKa = 11.84VEE1004 pKa = 3.59WSYY1007 pKa = 11.72SANRR1011 pKa = 11.84TLFAALAEE1019 pKa = 4.48GEE1021 pKa = 4.69SAFEE1025 pKa = 4.2TFTVTVDD1032 pKa = 3.64DD1033 pKa = 3.96GHH1035 pKa = 6.68GGSFSHH1041 pKa = 7.17DD1042 pKa = 3.24VVIEE1046 pKa = 4.03VQGVNDD1052 pKa = 3.68VPVAVADD1059 pKa = 3.98SLSTAEE1065 pKa = 4.48DD1066 pKa = 4.0TPLVIAAADD1075 pKa = 3.93LLANDD1080 pKa = 4.58SDD1082 pKa = 4.26VDD1084 pKa = 3.91GDD1086 pKa = 4.03TLTLVSVGGAVGGSVSLDD1104 pKa = 2.86AGQITFTPYY1113 pKa = 10.44TDD1115 pKa = 3.93FNGAASFAYY1124 pKa = 9.48MIADD1128 pKa = 3.85GKK1130 pKa = 11.29GGFDD1134 pKa = 3.35SATVAVTFTPVNDD1147 pKa = 3.67APVVGGQTSGAVVEE1161 pKa = 4.93DD1162 pKa = 3.94RR1163 pKa = 11.84TTSVSGALTIADD1175 pKa = 4.34LDD1177 pKa = 4.05AGEE1180 pKa = 5.02SAFQAQSGIAGTYY1193 pKa = 10.47GMLNINEE1200 pKa = 4.2QGQWTYY1206 pKa = 11.02FLSNASQAVQMLRR1219 pKa = 11.84GDD1221 pKa = 4.46GYY1223 pKa = 9.43PWSDD1227 pKa = 3.05NFTVLTADD1235 pKa = 3.57GTEE1238 pKa = 3.91QTISISVLGADD1249 pKa = 3.9DD1250 pKa = 4.3TVTATAPTTYY1260 pKa = 9.48TASTFTTLPVYY1271 pKa = 10.85ASGITVDD1278 pKa = 3.9SEE1280 pKa = 4.05TGYY1283 pKa = 11.4VYY1285 pKa = 10.66VAEE1288 pKa = 4.48KK1289 pKa = 10.98APVSKK1294 pKa = 10.41LYY1296 pKa = 10.56KK1297 pKa = 9.2VAPNGAVTQIASDD1310 pKa = 3.57FTGYY1314 pKa = 9.19TSGMGGYY1321 pKa = 10.3CYY1323 pKa = 10.05GTGDD1327 pKa = 3.27IEE1329 pKa = 4.75YY1330 pKa = 9.73YY1331 pKa = 10.25DD1332 pKa = 3.35GKK1334 pKa = 11.28VYY1336 pKa = 10.99VVTDD1340 pKa = 3.56AGKK1343 pKa = 10.47LVGIDD1348 pKa = 3.29VATGARR1354 pKa = 11.84SVLFTLPGGAGGPVLDD1370 pKa = 4.95AGIAITPDD1378 pKa = 3.14GKK1380 pKa = 11.17VLMSNGQSAPAGVWSYY1396 pKa = 11.36DD1397 pKa = 3.09IEE1399 pKa = 4.46TGASGFVASTPGATFEE1415 pKa = 4.72VDD1417 pKa = 5.02FDD1419 pKa = 4.04PATNKK1424 pKa = 9.23TVVGWPQNAVLTVNYY1439 pKa = 10.59AEE1441 pKa = 5.6DD1442 pKa = 3.29STGDD1446 pKa = 3.45YY1447 pKa = 10.4RR1448 pKa = 11.84GVGTPFYY1455 pKa = 10.99NFALSPGGDD1464 pKa = 3.51EE1465 pKa = 4.06IYY1467 pKa = 10.78LRR1469 pKa = 11.84SGSSLVQVNLGTGSSALVGSGLFNDD1494 pKa = 4.36SSQDD1498 pKa = 3.62LEE1500 pKa = 4.57FGPSSSGGGYY1510 pKa = 9.71SLYY1513 pKa = 10.47VVNVSTILEE1522 pKa = 4.32FKK1524 pKa = 11.03GFTTEE1529 pKa = 4.01WAQLPQDD1536 pKa = 4.52PDD1538 pKa = 4.77PIVLDD1543 pKa = 3.83LDD1545 pKa = 3.86GDD1547 pKa = 4.02GTYY1550 pKa = 10.55FSAPGTGVSFDD1561 pKa = 4.18LDD1563 pKa = 3.5GDD1565 pKa = 4.05GTAEE1569 pKa = 4.05LLGWASPADD1578 pKa = 3.63GVLVMDD1584 pKa = 5.51LDD1586 pKa = 4.22GSGAIEE1592 pKa = 4.46NGTEE1596 pKa = 4.19VVSEE1600 pKa = 4.05AFKK1603 pKa = 11.3GFGFGSSVDD1612 pKa = 3.56ALRR1615 pKa = 11.84SLDD1618 pKa = 3.75TNLDD1622 pKa = 3.44GMLDD1626 pKa = 3.5AGDD1629 pKa = 3.96AAFADD1634 pKa = 3.72LRR1636 pKa = 11.84LWRR1639 pKa = 11.84DD1640 pKa = 3.22GNGDD1644 pKa = 3.55GGSQGDD1650 pKa = 4.11EE1651 pKa = 4.19LLTLANLGITAISVEE1666 pKa = 3.95AQARR1670 pKa = 11.84DD1671 pKa = 3.65EE1672 pKa = 4.5TVDD1675 pKa = 3.34GHH1677 pKa = 6.36HH1678 pKa = 6.94VYY1680 pKa = 10.6AAGSFSYY1687 pKa = 11.05ASGATGSYY1695 pKa = 9.55VGARR1699 pKa = 11.84FAPASAGATDD1709 pKa = 4.97LEE1711 pKa = 4.9SGDD1714 pKa = 3.75EE1715 pKa = 4.37TEE1717 pKa = 5.45VIAEE1721 pKa = 4.52LFSAEE1726 pKa = 4.14DD1727 pKa = 3.89PEE1729 pKa = 4.42MEE1731 pKa = 4.51AALASALGASAPSGLRR1747 pKa = 11.84STRR1750 pKa = 11.84ADD1752 pKa = 3.36ALLALQDD1759 pKa = 5.82FFDD1762 pKa = 4.84DD1763 pKa = 3.63WQSFPGTSPAPADD1776 pKa = 3.38LRR1778 pKa = 11.84AAIGQWIGEE1787 pKa = 4.43VAFGDD1792 pKa = 3.76HH1793 pKa = 6.73SPTVDD1798 pKa = 3.83DD1799 pKa = 4.03LLHH1802 pKa = 7.05RR1803 pKa = 11.84FGHH1806 pKa = 6.8DD1807 pKa = 3.09PAFGNLLQGAHH1818 pKa = 6.43ASVNDD1823 pKa = 3.58MFIVV1827 pKa = 3.58
TT1 pKa = 6.13TAEE4 pKa = 4.26DD5 pKa = 4.01TPLVIAAADD14 pKa = 4.02LLANDD19 pKa = 4.79ADD21 pKa = 4.04IDD23 pKa = 4.24GDD25 pKa = 4.12TLSVTGVGGAVGGTVALDD43 pKa = 3.73DD44 pKa = 4.5GEE46 pKa = 4.52ITFTPDD52 pKa = 3.01ADD54 pKa = 4.24FNGDD58 pKa = 3.07ASFTYY63 pKa = 9.55TVSDD67 pKa = 3.78GKK69 pKa = 11.16GGTDD73 pKa = 2.94TATVNVTVTPVNDD86 pKa = 3.85APVAAADD93 pKa = 3.79TATTAEE99 pKa = 4.31DD100 pKa = 4.04TPLVIAAADD109 pKa = 4.02LLANDD114 pKa = 4.79ADD116 pKa = 4.04IDD118 pKa = 4.24GDD120 pKa = 4.12TLSVTGVGGAVGGTVALDD138 pKa = 3.83DD139 pKa = 4.66GEE141 pKa = 4.76VTFTPDD147 pKa = 3.42ANFNGDD153 pKa = 3.04ASFTYY158 pKa = 9.55TVSDD162 pKa = 3.78GKK164 pKa = 11.16GGTDD168 pKa = 2.94TATVNVTVTPVNDD181 pKa = 3.85APVAAADD188 pKa = 3.79TATTAEE194 pKa = 4.31DD195 pKa = 4.04TPLVIAAADD204 pKa = 4.02LLANDD209 pKa = 4.79ADD211 pKa = 4.04IDD213 pKa = 4.24GDD215 pKa = 4.12TLSVTGVGGAVGGTVALDD233 pKa = 3.83DD234 pKa = 4.66GEE236 pKa = 4.76VTFTPDD242 pKa = 3.42ANFNGDD248 pKa = 3.04ASFTYY253 pKa = 9.55TVSDD257 pKa = 3.78GKK259 pKa = 11.16GGTDD263 pKa = 2.94TATVNVTVTPVNDD276 pKa = 3.74APIATADD283 pKa = 3.57TATTAEE289 pKa = 4.31DD290 pKa = 4.14TPLVIAVADD299 pKa = 4.29LLANDD304 pKa = 4.6SDD306 pKa = 5.16LDD308 pKa = 4.06GDD310 pKa = 4.22TLSVTGVGGAVGGTVALDD328 pKa = 3.73DD329 pKa = 4.5GEE331 pKa = 4.52ITFTPDD337 pKa = 3.01ADD339 pKa = 4.24FNGDD343 pKa = 3.01ASFSYY348 pKa = 9.9TVSDD352 pKa = 3.88GKK354 pKa = 11.18GGTDD358 pKa = 3.11TATVGVTVTPVNDD371 pKa = 3.73APVVASGATATFAEE385 pKa = 4.62NGTGAVYY392 pKa = 10.33QIAASDD398 pKa = 4.07AEE400 pKa = 4.67DD401 pKa = 5.25DD402 pKa = 4.09GLTYY406 pKa = 11.06ALSGADD412 pKa = 3.29ADD414 pKa = 4.86LFTVDD419 pKa = 4.79ADD421 pKa = 3.69GKK423 pKa = 9.95VAFKK427 pKa = 10.52AAPDD431 pKa = 3.63VEE433 pKa = 4.77APADD437 pKa = 3.64AGGDD441 pKa = 3.45NVYY444 pKa = 11.09DD445 pKa = 3.74LVVEE449 pKa = 4.43VSDD452 pKa = 4.56GTDD455 pKa = 3.16TTSKK459 pKa = 10.78AVAITVTDD467 pKa = 3.61VAEE470 pKa = 4.31NSAPRR475 pKa = 11.84LVSAPVLGGITAEE488 pKa = 4.02AAAVGNGFSSSARR501 pKa = 11.84FSPNGTKK508 pKa = 10.34VLFQSTASNLVEE520 pKa = 4.24GDD522 pKa = 3.52ANGTEE527 pKa = 4.31DD528 pKa = 4.81VFVKK532 pKa = 10.6DD533 pKa = 4.38LEE535 pKa = 4.63TGAVTLVSSSSSGTPGNGWSNGHH558 pKa = 5.75QFSADD563 pKa = 3.1GTKK566 pKa = 10.55VLFYY570 pKa = 10.68SYY572 pKa = 11.27ASNLVAGDD580 pKa = 3.5INGAADD586 pKa = 3.86VFVKK590 pKa = 10.61DD591 pKa = 4.39LEE593 pKa = 4.63TGAVTLVSSSSSGTPGNGEE612 pKa = 3.66SSGYY616 pKa = 9.64QFSADD621 pKa = 3.2GTKK624 pKa = 10.55VLFYY628 pKa = 10.83SYY630 pKa = 11.43ANNLVAGDD638 pKa = 3.84INGVADD644 pKa = 3.75VFVKK648 pKa = 10.69NLEE651 pKa = 4.25TGAVTLVSSSSSGTPGNGTSSGYY674 pKa = 9.85QFSADD679 pKa = 3.2GTKK682 pKa = 10.55VLFYY686 pKa = 10.83SYY688 pKa = 11.43ANNLVAGDD696 pKa = 3.84INGVADD702 pKa = 3.75VFVKK706 pKa = 10.69NLEE709 pKa = 4.25TGAVTLVSSSSSGTPGNGTSYY730 pKa = 10.69GHH732 pKa = 6.03QFSADD737 pKa = 3.18GTKK740 pKa = 10.55VLFYY744 pKa = 10.83SYY746 pKa = 11.43ANNLVAGDD754 pKa = 3.84INGVADD760 pKa = 3.75VFVKK764 pKa = 10.69NLEE767 pKa = 4.25TGAVTLVSSSSSGTPGNGTSYY788 pKa = 10.7GYY790 pKa = 10.37QYY792 pKa = 11.09SADD795 pKa = 3.58GTKK798 pKa = 10.58VLFQSDD804 pKa = 3.69ASNLVAGDD812 pKa = 3.5INGAADD818 pKa = 3.69IFVKK822 pKa = 10.61DD823 pKa = 4.06LEE825 pKa = 4.72TGVVTLVSSSSSGTPGNQTSYY846 pKa = 10.27SHH848 pKa = 6.19QFSADD853 pKa = 3.09GTKK856 pKa = 10.47VLFYY860 pKa = 10.89SSASNLVAGDD870 pKa = 3.5INGAADD876 pKa = 3.86VFVKK880 pKa = 10.53DD881 pKa = 4.33LEE883 pKa = 4.74SGAVTLVSSSSSGTPGNGTSYY904 pKa = 10.69GHH906 pKa = 6.21QFSADD911 pKa = 3.35CAKK914 pKa = 10.82VLFRR918 pKa = 11.84SYY920 pKa = 11.39ASNLVAGDD928 pKa = 3.6TNGTYY933 pKa = 10.75DD934 pKa = 4.52VFMKK938 pKa = 10.72DD939 pKa = 3.12LATGEE944 pKa = 4.33VTLLSARR951 pKa = 11.84SLAATPLAKK960 pKa = 10.31SGAFTFDD967 pKa = 3.91DD968 pKa = 4.07ADD970 pKa = 4.01GGDD973 pKa = 3.38THH975 pKa = 6.5TASVLPAAGTLGTLQADD992 pKa = 3.52IVVVADD998 pKa = 3.94GPDD1001 pKa = 3.29RR1002 pKa = 11.84VEE1004 pKa = 3.59WSYY1007 pKa = 11.72SANRR1011 pKa = 11.84TLFAALAEE1019 pKa = 4.48GEE1021 pKa = 4.69SAFEE1025 pKa = 4.2TFTVTVDD1032 pKa = 3.64DD1033 pKa = 3.96GHH1035 pKa = 6.68GGSFSHH1041 pKa = 7.17DD1042 pKa = 3.24VVIEE1046 pKa = 4.03VQGVNDD1052 pKa = 3.68VPVAVADD1059 pKa = 3.98SLSTAEE1065 pKa = 4.48DD1066 pKa = 4.0TPLVIAAADD1075 pKa = 3.93LLANDD1080 pKa = 4.58SDD1082 pKa = 4.26VDD1084 pKa = 3.91GDD1086 pKa = 4.03TLTLVSVGGAVGGSVSLDD1104 pKa = 2.86AGQITFTPYY1113 pKa = 10.44TDD1115 pKa = 3.93FNGAASFAYY1124 pKa = 9.48MIADD1128 pKa = 3.85GKK1130 pKa = 11.29GGFDD1134 pKa = 3.35SATVAVTFTPVNDD1147 pKa = 3.67APVVGGQTSGAVVEE1161 pKa = 4.93DD1162 pKa = 3.94RR1163 pKa = 11.84TTSVSGALTIADD1175 pKa = 4.34LDD1177 pKa = 4.05AGEE1180 pKa = 5.02SAFQAQSGIAGTYY1193 pKa = 10.47GMLNINEE1200 pKa = 4.2QGQWTYY1206 pKa = 11.02FLSNASQAVQMLRR1219 pKa = 11.84GDD1221 pKa = 4.46GYY1223 pKa = 9.43PWSDD1227 pKa = 3.05NFTVLTADD1235 pKa = 3.57GTEE1238 pKa = 3.91QTISISVLGADD1249 pKa = 3.9DD1250 pKa = 4.3TVTATAPTTYY1260 pKa = 9.48TASTFTTLPVYY1271 pKa = 10.85ASGITVDD1278 pKa = 3.9SEE1280 pKa = 4.05TGYY1283 pKa = 11.4VYY1285 pKa = 10.66VAEE1288 pKa = 4.48KK1289 pKa = 10.98APVSKK1294 pKa = 10.41LYY1296 pKa = 10.56KK1297 pKa = 9.2VAPNGAVTQIASDD1310 pKa = 3.57FTGYY1314 pKa = 9.19TSGMGGYY1321 pKa = 10.3CYY1323 pKa = 10.05GTGDD1327 pKa = 3.27IEE1329 pKa = 4.75YY1330 pKa = 9.73YY1331 pKa = 10.25DD1332 pKa = 3.35GKK1334 pKa = 11.28VYY1336 pKa = 10.99VVTDD1340 pKa = 3.56AGKK1343 pKa = 10.47LVGIDD1348 pKa = 3.29VATGARR1354 pKa = 11.84SVLFTLPGGAGGPVLDD1370 pKa = 4.95AGIAITPDD1378 pKa = 3.14GKK1380 pKa = 11.17VLMSNGQSAPAGVWSYY1396 pKa = 11.36DD1397 pKa = 3.09IEE1399 pKa = 4.46TGASGFVASTPGATFEE1415 pKa = 4.72VDD1417 pKa = 5.02FDD1419 pKa = 4.04PATNKK1424 pKa = 9.23TVVGWPQNAVLTVNYY1439 pKa = 10.59AEE1441 pKa = 5.6DD1442 pKa = 3.29STGDD1446 pKa = 3.45YY1447 pKa = 10.4RR1448 pKa = 11.84GVGTPFYY1455 pKa = 10.99NFALSPGGDD1464 pKa = 3.51EE1465 pKa = 4.06IYY1467 pKa = 10.78LRR1469 pKa = 11.84SGSSLVQVNLGTGSSALVGSGLFNDD1494 pKa = 4.36SSQDD1498 pKa = 3.62LEE1500 pKa = 4.57FGPSSSGGGYY1510 pKa = 9.71SLYY1513 pKa = 10.47VVNVSTILEE1522 pKa = 4.32FKK1524 pKa = 11.03GFTTEE1529 pKa = 4.01WAQLPQDD1536 pKa = 4.52PDD1538 pKa = 4.77PIVLDD1543 pKa = 3.83LDD1545 pKa = 3.86GDD1547 pKa = 4.02GTYY1550 pKa = 10.55FSAPGTGVSFDD1561 pKa = 4.18LDD1563 pKa = 3.5GDD1565 pKa = 4.05GTAEE1569 pKa = 4.05LLGWASPADD1578 pKa = 3.63GVLVMDD1584 pKa = 5.51LDD1586 pKa = 4.22GSGAIEE1592 pKa = 4.46NGTEE1596 pKa = 4.19VVSEE1600 pKa = 4.05AFKK1603 pKa = 11.3GFGFGSSVDD1612 pKa = 3.56ALRR1615 pKa = 11.84SLDD1618 pKa = 3.75TNLDD1622 pKa = 3.44GMLDD1626 pKa = 3.5AGDD1629 pKa = 3.96AAFADD1634 pKa = 3.72LRR1636 pKa = 11.84LWRR1639 pKa = 11.84DD1640 pKa = 3.22GNGDD1644 pKa = 3.55GGSQGDD1650 pKa = 4.11EE1651 pKa = 4.19LLTLANLGITAISVEE1666 pKa = 3.95AQARR1670 pKa = 11.84DD1671 pKa = 3.65EE1672 pKa = 4.5TVDD1675 pKa = 3.34GHH1677 pKa = 6.36HH1678 pKa = 6.94VYY1680 pKa = 10.6AAGSFSYY1687 pKa = 11.05ASGATGSYY1695 pKa = 9.55VGARR1699 pKa = 11.84FAPASAGATDD1709 pKa = 4.97LEE1711 pKa = 4.9SGDD1714 pKa = 3.75EE1715 pKa = 4.37TEE1717 pKa = 5.45VIAEE1721 pKa = 4.52LFSAEE1726 pKa = 4.14DD1727 pKa = 3.89PEE1729 pKa = 4.42MEE1731 pKa = 4.51AALASALGASAPSGLRR1747 pKa = 11.84STRR1750 pKa = 11.84ADD1752 pKa = 3.36ALLALQDD1759 pKa = 5.82FFDD1762 pKa = 4.84DD1763 pKa = 3.63WQSFPGTSPAPADD1776 pKa = 3.38LRR1778 pKa = 11.84AAIGQWIGEE1787 pKa = 4.43VAFGDD1792 pKa = 3.76HH1793 pKa = 6.73SPTVDD1798 pKa = 3.83DD1799 pKa = 4.03LLHH1802 pKa = 7.05RR1803 pKa = 11.84FGHH1806 pKa = 6.8DD1807 pKa = 3.09PAFGNLLQGAHH1818 pKa = 6.43ASVNDD1823 pKa = 3.58MFIVV1827 pKa = 3.58
Molecular weight: 183.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3L7ANG0|A0A3L7ANG0_9RHIZ Cobalt-precorrin-5B C(1)-methyltransferase OS=Xanthobacter tagetidis OX=60216 GN=cbiD PE=3 SV=1
MM1 pKa = 7.48PRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84SRR7 pKa = 11.84SRR9 pKa = 11.84RR10 pKa = 11.84SPWPAPRR17 pKa = 11.84RR18 pKa = 11.84SPTRR22 pKa = 11.84PRR24 pKa = 11.84PTSARR29 pKa = 11.84PASMRR34 pKa = 11.84RR35 pKa = 11.84PRR37 pKa = 11.84PFTRR41 pKa = 11.84PPHH44 pKa = 5.94LASPPRR50 pKa = 11.84RR51 pKa = 11.84TSAASSPTRR60 pKa = 11.84CAGWW64 pKa = 3.2
MM1 pKa = 7.48PRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84SRR7 pKa = 11.84SRR9 pKa = 11.84RR10 pKa = 11.84SPWPAPRR17 pKa = 11.84RR18 pKa = 11.84SPTRR22 pKa = 11.84PRR24 pKa = 11.84PTSARR29 pKa = 11.84PASMRR34 pKa = 11.84RR35 pKa = 11.84PRR37 pKa = 11.84PFTRR41 pKa = 11.84PPHH44 pKa = 5.94LASPPRR50 pKa = 11.84RR51 pKa = 11.84TSAASSPTRR60 pKa = 11.84CAGWW64 pKa = 3.2
Molecular weight: 7.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1423151 |
29 |
3175 |
320.4 |
34.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.58 ± 0.064 | 0.862 ± 0.012 |
5.376 ± 0.031 | 5.413 ± 0.033 |
3.694 ± 0.025 | 9.091 ± 0.06 |
1.919 ± 0.019 | 4.565 ± 0.029 |
2.944 ± 0.029 | 10.358 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.373 ± 0.016 | 2.089 ± 0.022 |
5.797 ± 0.038 | 2.707 ± 0.019 |
7.327 ± 0.043 | 4.79 ± 0.03 |
4.954 ± 0.029 | 7.919 ± 0.028 |
1.203 ± 0.014 | 2.039 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
