
Sphingomonas gilva
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
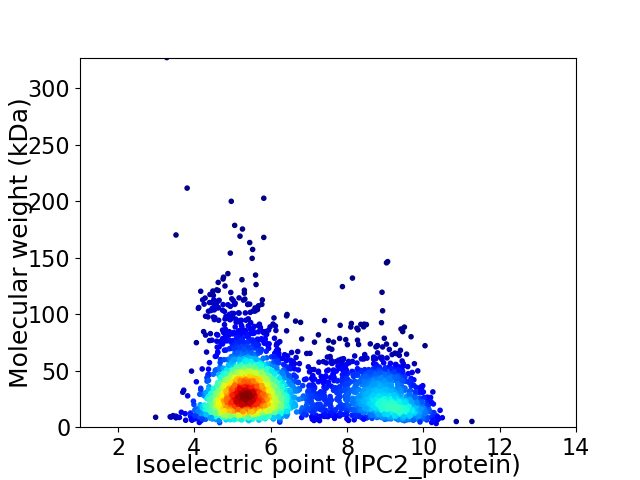
Virtual 2D-PAGE plot for 3257 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A396RSC3|A0A396RSC3_9SPHN Arylsulfatase OS=Sphingomonas gilva OX=2305907 GN=D1610_04180 PE=3 SV=1
MM1 pKa = 7.61AATPALAQDD10 pKa = 3.81TQDD13 pKa = 3.44PSVAPEE19 pKa = 3.89TQTAEE24 pKa = 4.41DD25 pKa = 4.59ADD27 pKa = 3.88TGEE30 pKa = 4.76NIVVTGSILRR40 pKa = 11.84RR41 pKa = 11.84STADD45 pKa = 2.98TVSPITVVTADD56 pKa = 4.14DD57 pKa = 3.88LDD59 pKa = 3.96ARR61 pKa = 11.84GINTTQEE68 pKa = 4.19AIQRR72 pKa = 11.84LSSNNGPALTNSFSANGAFAAGASAVSLRR101 pKa = 11.84GLSTSSTLVLFDD113 pKa = 3.59GMRR116 pKa = 11.84ASYY119 pKa = 10.86YY120 pKa = 10.12PLADD124 pKa = 3.79DD125 pKa = 3.57ATRR128 pKa = 11.84NFVDD132 pKa = 5.16LNTIPDD138 pKa = 4.74DD139 pKa = 3.19IVEE142 pKa = 4.21RR143 pKa = 11.84VEE145 pKa = 3.88VLRR148 pKa = 11.84DD149 pKa = 3.72GASSTYY155 pKa = 10.5GADD158 pKa = 3.69AIAGVVNVITKK169 pKa = 10.36RR170 pKa = 11.84EE171 pKa = 3.87ITGISARR178 pKa = 11.84AEE180 pKa = 3.78AGIDD184 pKa = 3.45EE185 pKa = 5.23RR186 pKa = 11.84GNSPNYY192 pKa = 9.84RR193 pKa = 11.84LSLTGGFGDD202 pKa = 4.93VEE204 pKa = 4.25GDD206 pKa = 3.64GYY208 pKa = 11.62NVYY211 pKa = 11.02ASGFYY216 pKa = 8.61YY217 pKa = 9.99TQDD220 pKa = 3.05ALFNGDD226 pKa = 4.51LPYY229 pKa = 10.48PYY231 pKa = 10.4NSQNQSGICLDD242 pKa = 4.44GVCGPQVGPNSGSINPSSQGFEE264 pKa = 4.23GFSLTSDD271 pKa = 2.82RR272 pKa = 11.84RR273 pKa = 11.84LFATTFFVAPYY284 pKa = 9.29DD285 pKa = 3.86ASNTTSQGRR294 pKa = 11.84YY295 pKa = 7.24QFLNPAAGCITGEE308 pKa = 4.12TPYY311 pKa = 10.04TLSAAEE317 pKa = 4.89FADD320 pKa = 4.07PDD322 pKa = 3.74NALSPRR328 pKa = 11.84TVCQGDD334 pKa = 4.2LVTQYY339 pKa = 11.44GVVSPEE345 pKa = 3.77LEE347 pKa = 4.07RR348 pKa = 11.84FGGAIKK354 pKa = 9.39GTIALGGGIEE364 pKa = 4.05ASLVGNYY371 pKa = 8.68LQSKK375 pKa = 9.09VSYY378 pKa = 8.8TGEE381 pKa = 3.98PATIRR386 pKa = 11.84ANANTGIMFPRR397 pKa = 11.84FSTFTGGPPNAPGSFALALPVYY419 pKa = 9.76VCPLVNGMPQATCDD433 pKa = 3.61ATNGTLNPNNPFAAQGQVARR453 pKa = 11.84IIGRR457 pKa = 11.84IPNLIEE463 pKa = 3.92YY464 pKa = 10.45NEE466 pKa = 4.36TLSRR470 pKa = 11.84TYY472 pKa = 10.53RR473 pKa = 11.84GALTISGPISDD484 pKa = 3.96NWNFAVDD491 pKa = 3.9AVGMRR496 pKa = 11.84TDD498 pKa = 3.89LRR500 pKa = 11.84RR501 pKa = 11.84TQEE504 pKa = 4.07GYY506 pKa = 11.26VYY508 pKa = 10.29IQRR511 pKa = 11.84LLNVVADD518 pKa = 4.26GSYY521 pKa = 11.19NFVDD525 pKa = 4.43PFANSQATLDD535 pKa = 3.91YY536 pKa = 10.83LSPDD540 pKa = 3.3NVTDD544 pKa = 3.58SHH546 pKa = 8.15SDD548 pKa = 2.89IYY550 pKa = 11.1AASANISGSLFEE562 pKa = 5.33LPGGPLQLGVGAAIRR577 pKa = 11.84NEE579 pKa = 4.44SIDD582 pKa = 3.8APSANDD588 pKa = 3.84DD589 pKa = 3.65FAGPTEE595 pKa = 4.42RR596 pKa = 11.84YY597 pKa = 8.86FVLNAFGTSGEE608 pKa = 4.17RR609 pKa = 11.84TVYY612 pKa = 10.63SAFAEE617 pKa = 3.91LDD619 pKa = 3.54APILEE624 pKa = 4.39WVDD627 pKa = 3.44VNLSGRR633 pKa = 11.84YY634 pKa = 9.37DD635 pKa = 3.64NYY637 pKa = 11.33SSGQDD642 pKa = 2.98AFSPKK647 pKa = 9.44VGVRR651 pKa = 11.84FRR653 pKa = 11.84PFDD656 pKa = 3.6GLTVRR661 pKa = 11.84GTWSRR666 pKa = 11.84GFRR669 pKa = 11.84IPSFAEE675 pKa = 3.8ANALPTTGFVTNTAALFNDD694 pKa = 4.64TYY696 pKa = 10.83LAQYY700 pKa = 9.2GCTVATFNACPTYY713 pKa = 10.46IRR715 pKa = 11.84AGAYY719 pKa = 8.88GQTTLASPDD728 pKa = 3.86LDD730 pKa = 3.95PEE732 pKa = 4.69RR733 pKa = 11.84STSWTAGIVFEE744 pKa = 4.36PLRR747 pKa = 11.84NVTLAVDD754 pKa = 3.71YY755 pKa = 11.44YY756 pKa = 10.99NIKK759 pKa = 8.7KK760 pKa = 8.47TGAITTPSNSPALIAYY776 pKa = 8.89YY777 pKa = 10.06SGQAIPEE784 pKa = 4.62GYY786 pKa = 10.27NVIADD791 pKa = 3.85APDD794 pKa = 3.39AQFPNATPRR803 pKa = 11.84VAFVQSSLINANTIRR818 pKa = 11.84SEE820 pKa = 4.13GLDD823 pKa = 3.5FAARR827 pKa = 11.84ASFDD831 pKa = 3.8LGPDD835 pKa = 2.89IRR837 pKa = 11.84FTSSAEE843 pKa = 3.31ASYY846 pKa = 10.45IINLSTEE853 pKa = 4.1FPDD856 pKa = 4.77GSVEE860 pKa = 4.32SYY862 pKa = 10.88EE863 pKa = 4.13GTLGNFNLTAGSGTPEE879 pKa = 3.73WHH881 pKa = 7.07GSWQNTLEE889 pKa = 4.02MGAWTLTATAEE900 pKa = 4.17YY901 pKa = 10.25FDD903 pKa = 4.97GYY905 pKa = 11.0NLSAEE910 pKa = 4.36DD911 pKa = 3.48QNGAGTSGDD920 pKa = 3.73CGLLSEE926 pKa = 5.46AFVEE930 pKa = 4.5CDD932 pKa = 3.1VPSYY936 pKa = 10.2VTVDD940 pKa = 3.69LSTSFDD946 pKa = 3.62VNDD949 pKa = 3.38NFTFYY954 pKa = 11.72VNVLNVFDD962 pKa = 5.03NLPPIDD968 pKa = 5.35PITYY972 pKa = 9.46GANNYY977 pKa = 10.07NPVQGGTGIFGRR989 pKa = 11.84SFRR992 pKa = 11.84AGAKK996 pKa = 10.22VNFF999 pKa = 4.21
MM1 pKa = 7.61AATPALAQDD10 pKa = 3.81TQDD13 pKa = 3.44PSVAPEE19 pKa = 3.89TQTAEE24 pKa = 4.41DD25 pKa = 4.59ADD27 pKa = 3.88TGEE30 pKa = 4.76NIVVTGSILRR40 pKa = 11.84RR41 pKa = 11.84STADD45 pKa = 2.98TVSPITVVTADD56 pKa = 4.14DD57 pKa = 3.88LDD59 pKa = 3.96ARR61 pKa = 11.84GINTTQEE68 pKa = 4.19AIQRR72 pKa = 11.84LSSNNGPALTNSFSANGAFAAGASAVSLRR101 pKa = 11.84GLSTSSTLVLFDD113 pKa = 3.59GMRR116 pKa = 11.84ASYY119 pKa = 10.86YY120 pKa = 10.12PLADD124 pKa = 3.79DD125 pKa = 3.57ATRR128 pKa = 11.84NFVDD132 pKa = 5.16LNTIPDD138 pKa = 4.74DD139 pKa = 3.19IVEE142 pKa = 4.21RR143 pKa = 11.84VEE145 pKa = 3.88VLRR148 pKa = 11.84DD149 pKa = 3.72GASSTYY155 pKa = 10.5GADD158 pKa = 3.69AIAGVVNVITKK169 pKa = 10.36RR170 pKa = 11.84EE171 pKa = 3.87ITGISARR178 pKa = 11.84AEE180 pKa = 3.78AGIDD184 pKa = 3.45EE185 pKa = 5.23RR186 pKa = 11.84GNSPNYY192 pKa = 9.84RR193 pKa = 11.84LSLTGGFGDD202 pKa = 4.93VEE204 pKa = 4.25GDD206 pKa = 3.64GYY208 pKa = 11.62NVYY211 pKa = 11.02ASGFYY216 pKa = 8.61YY217 pKa = 9.99TQDD220 pKa = 3.05ALFNGDD226 pKa = 4.51LPYY229 pKa = 10.48PYY231 pKa = 10.4NSQNQSGICLDD242 pKa = 4.44GVCGPQVGPNSGSINPSSQGFEE264 pKa = 4.23GFSLTSDD271 pKa = 2.82RR272 pKa = 11.84RR273 pKa = 11.84LFATTFFVAPYY284 pKa = 9.29DD285 pKa = 3.86ASNTTSQGRR294 pKa = 11.84YY295 pKa = 7.24QFLNPAAGCITGEE308 pKa = 4.12TPYY311 pKa = 10.04TLSAAEE317 pKa = 4.89FADD320 pKa = 4.07PDD322 pKa = 3.74NALSPRR328 pKa = 11.84TVCQGDD334 pKa = 4.2LVTQYY339 pKa = 11.44GVVSPEE345 pKa = 3.77LEE347 pKa = 4.07RR348 pKa = 11.84FGGAIKK354 pKa = 9.39GTIALGGGIEE364 pKa = 4.05ASLVGNYY371 pKa = 8.68LQSKK375 pKa = 9.09VSYY378 pKa = 8.8TGEE381 pKa = 3.98PATIRR386 pKa = 11.84ANANTGIMFPRR397 pKa = 11.84FSTFTGGPPNAPGSFALALPVYY419 pKa = 9.76VCPLVNGMPQATCDD433 pKa = 3.61ATNGTLNPNNPFAAQGQVARR453 pKa = 11.84IIGRR457 pKa = 11.84IPNLIEE463 pKa = 3.92YY464 pKa = 10.45NEE466 pKa = 4.36TLSRR470 pKa = 11.84TYY472 pKa = 10.53RR473 pKa = 11.84GALTISGPISDD484 pKa = 3.96NWNFAVDD491 pKa = 3.9AVGMRR496 pKa = 11.84TDD498 pKa = 3.89LRR500 pKa = 11.84RR501 pKa = 11.84TQEE504 pKa = 4.07GYY506 pKa = 11.26VYY508 pKa = 10.29IQRR511 pKa = 11.84LLNVVADD518 pKa = 4.26GSYY521 pKa = 11.19NFVDD525 pKa = 4.43PFANSQATLDD535 pKa = 3.91YY536 pKa = 10.83LSPDD540 pKa = 3.3NVTDD544 pKa = 3.58SHH546 pKa = 8.15SDD548 pKa = 2.89IYY550 pKa = 11.1AASANISGSLFEE562 pKa = 5.33LPGGPLQLGVGAAIRR577 pKa = 11.84NEE579 pKa = 4.44SIDD582 pKa = 3.8APSANDD588 pKa = 3.84DD589 pKa = 3.65FAGPTEE595 pKa = 4.42RR596 pKa = 11.84YY597 pKa = 8.86FVLNAFGTSGEE608 pKa = 4.17RR609 pKa = 11.84TVYY612 pKa = 10.63SAFAEE617 pKa = 3.91LDD619 pKa = 3.54APILEE624 pKa = 4.39WVDD627 pKa = 3.44VNLSGRR633 pKa = 11.84YY634 pKa = 9.37DD635 pKa = 3.64NYY637 pKa = 11.33SSGQDD642 pKa = 2.98AFSPKK647 pKa = 9.44VGVRR651 pKa = 11.84FRR653 pKa = 11.84PFDD656 pKa = 3.6GLTVRR661 pKa = 11.84GTWSRR666 pKa = 11.84GFRR669 pKa = 11.84IPSFAEE675 pKa = 3.8ANALPTTGFVTNTAALFNDD694 pKa = 4.64TYY696 pKa = 10.83LAQYY700 pKa = 9.2GCTVATFNACPTYY713 pKa = 10.46IRR715 pKa = 11.84AGAYY719 pKa = 8.88GQTTLASPDD728 pKa = 3.86LDD730 pKa = 3.95PEE732 pKa = 4.69RR733 pKa = 11.84STSWTAGIVFEE744 pKa = 4.36PLRR747 pKa = 11.84NVTLAVDD754 pKa = 3.71YY755 pKa = 11.44YY756 pKa = 10.99NIKK759 pKa = 8.7KK760 pKa = 8.47TGAITTPSNSPALIAYY776 pKa = 8.89YY777 pKa = 10.06SGQAIPEE784 pKa = 4.62GYY786 pKa = 10.27NVIADD791 pKa = 3.85APDD794 pKa = 3.39AQFPNATPRR803 pKa = 11.84VAFVQSSLINANTIRR818 pKa = 11.84SEE820 pKa = 4.13GLDD823 pKa = 3.5FAARR827 pKa = 11.84ASFDD831 pKa = 3.8LGPDD835 pKa = 2.89IRR837 pKa = 11.84FTSSAEE843 pKa = 3.31ASYY846 pKa = 10.45IINLSTEE853 pKa = 4.1FPDD856 pKa = 4.77GSVEE860 pKa = 4.32SYY862 pKa = 10.88EE863 pKa = 4.13GTLGNFNLTAGSGTPEE879 pKa = 3.73WHH881 pKa = 7.07GSWQNTLEE889 pKa = 4.02MGAWTLTATAEE900 pKa = 4.17YY901 pKa = 10.25FDD903 pKa = 4.97GYY905 pKa = 11.0NLSAEE910 pKa = 4.36DD911 pKa = 3.48QNGAGTSGDD920 pKa = 3.73CGLLSEE926 pKa = 5.46AFVEE930 pKa = 4.5CDD932 pKa = 3.1VPSYY936 pKa = 10.2VTVDD940 pKa = 3.69LSTSFDD946 pKa = 3.62VNDD949 pKa = 3.38NFTFYY954 pKa = 11.72VNVLNVFDD962 pKa = 5.03NLPPIDD968 pKa = 5.35PITYY972 pKa = 9.46GANNYY977 pKa = 10.07NPVQGGTGIFGRR989 pKa = 11.84SFRR992 pKa = 11.84AGAKK996 pKa = 10.22VNFF999 pKa = 4.21
Molecular weight: 105.97 kDa
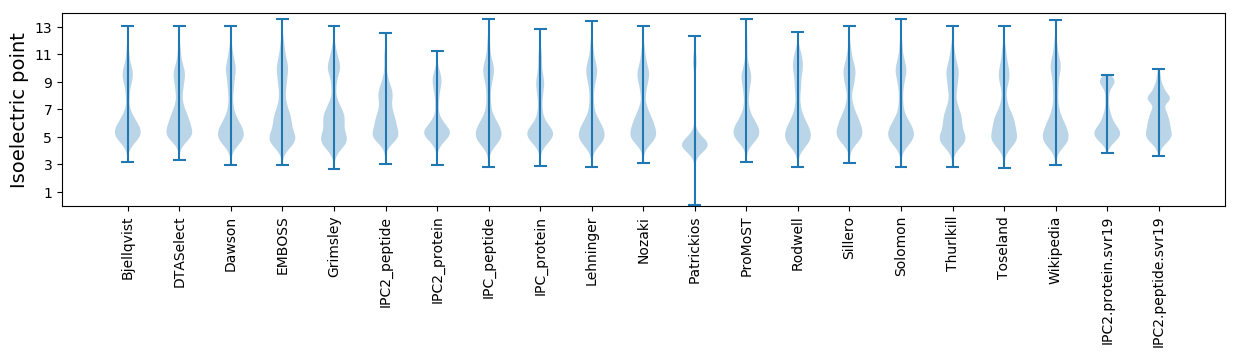
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A396RRU7|A0A396RRU7_9SPHN p-hydroxycinnamoyl CoA hydratase/lyase OS=Sphingomonas gilva OX=2305907 GN=D1610_03945 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84SRR21 pKa = 11.84MATPGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84SRR21 pKa = 11.84MATPGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1067601 |
30 |
3520 |
327.8 |
35.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.045 ± 0.073 | 0.695 ± 0.012 |
6.145 ± 0.035 | 5.574 ± 0.042 |
3.498 ± 0.029 | 9.225 ± 0.052 |
1.896 ± 0.025 | 4.943 ± 0.029 |
2.666 ± 0.038 | 9.595 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.331 ± 0.025 | 2.422 ± 0.032 |
5.463 ± 0.034 | 2.879 ± 0.024 |
7.755 ± 0.052 | 4.85 ± 0.038 |
5.218 ± 0.047 | 7.12 ± 0.036 |
1.469 ± 0.019 | 2.211 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
