
Circovirus-like genome CB-A
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.77
Get precalculated fractions of proteins
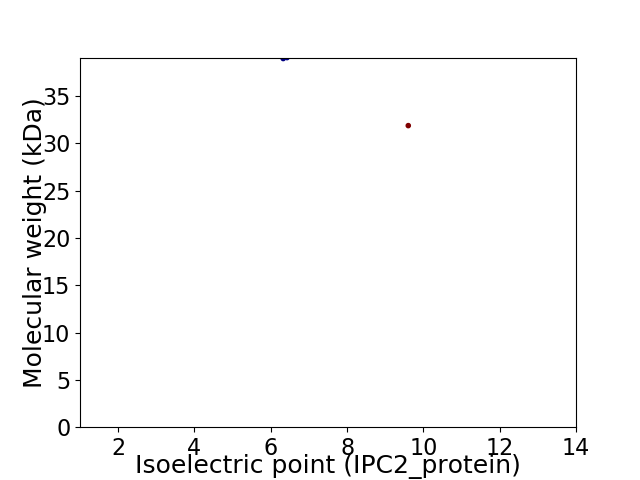
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6GII6|C6GII6_9VIRU Uncharacterized protein OS=Circovirus-like genome CB-A OX=642256 PE=4 SV=1
MM1 pKa = 7.17SRR3 pKa = 11.84SRR5 pKa = 11.84NWCFTYY11 pKa = 10.93NNYY14 pKa = 10.46DD15 pKa = 3.28SSPRR19 pKa = 11.84IQKK22 pKa = 9.5CMKK25 pKa = 8.58YY26 pKa = 8.75LTYY29 pKa = 10.05GYY31 pKa = 10.22EE32 pKa = 4.13VGEE35 pKa = 4.38SGTPHH40 pKa = 6.23LQGFVIFKK48 pKa = 10.37NAVAKK53 pKa = 10.07PSQYY57 pKa = 10.52FKK59 pKa = 10.39PAYY62 pKa = 9.51HH63 pKa = 6.33FEE65 pKa = 4.33KK66 pKa = 10.88ARR68 pKa = 11.84GTPQQALEE76 pKa = 4.09YY77 pKa = 9.06CQKK80 pKa = 10.96DD81 pKa = 3.74GNFKK85 pKa = 10.9EE86 pKa = 5.01FGEE89 pKa = 4.41KK90 pKa = 10.06PKK92 pKa = 10.42TVADD96 pKa = 4.08GGAAGGEE103 pKa = 4.23ANKK106 pKa = 10.03RR107 pKa = 11.84RR108 pKa = 11.84YY109 pKa = 9.66EE110 pKa = 4.1EE111 pKa = 4.15AFSAAKK117 pKa = 9.47EE118 pKa = 3.99GRR120 pKa = 11.84MDD122 pKa = 6.65DD123 pKa = 3.7IPADD127 pKa = 3.17IYY129 pKa = 10.48IRR131 pKa = 11.84HH132 pKa = 6.06YY133 pKa = 10.24STLKK137 pKa = 10.52KK138 pKa = 10.1IRR140 pKa = 11.84FDD142 pKa = 3.67HH143 pKa = 7.29APPAQNNDD151 pKa = 2.93VLNNYY156 pKa = 7.25WVYY159 pKa = 11.03GPSGTGKK166 pKa = 9.65SKK168 pKa = 10.42SVRR171 pKa = 11.84EE172 pKa = 4.07FFGKK176 pKa = 9.85SLYY179 pKa = 10.56VKK181 pKa = 10.22NQNKK185 pKa = 8.09WFDD188 pKa = 3.68GYY190 pKa = 10.93EE191 pKa = 4.21GEE193 pKa = 5.11DD194 pKa = 3.53FVLIDD199 pKa = 4.53DD200 pKa = 4.5VHH202 pKa = 6.64PNWSGKK208 pKa = 8.88TILKK212 pKa = 9.42IWSDD216 pKa = 4.05HH217 pKa = 5.82YY218 pKa = 11.05PFSPEE223 pKa = 3.73TKK225 pKa = 9.5GGHH228 pKa = 5.51IKK230 pKa = 10.07MIRR233 pKa = 11.84PEE235 pKa = 4.47GIIVTSNYY243 pKa = 9.41TIEE246 pKa = 4.15EE247 pKa = 4.21MYY249 pKa = 10.47EE250 pKa = 4.0AEE252 pKa = 5.16EE253 pKa = 4.07DD254 pKa = 3.51RR255 pKa = 11.84QPIRR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84FKK263 pKa = 10.39VIKK266 pKa = 10.46SEE268 pKa = 3.89AGYY271 pKa = 10.7DD272 pKa = 3.83VIMKK276 pKa = 9.25EE277 pKa = 3.94YY278 pKa = 9.48NKK280 pKa = 10.19RR281 pKa = 11.84VAATKK286 pKa = 9.79ICSNTDD292 pKa = 2.73EE293 pKa = 4.6DD294 pKa = 4.71VVVAEE299 pKa = 5.19HH300 pKa = 6.42EE301 pKa = 4.23PCRR304 pKa = 11.84PATRR308 pKa = 11.84ASAPGDD314 pKa = 3.63HH315 pKa = 6.94EE316 pKa = 4.89LPPPATEE323 pKa = 3.89VQSIVVAVIEE333 pKa = 4.74SPPPPATTDD342 pKa = 3.12QQ343 pKa = 3.5
MM1 pKa = 7.17SRR3 pKa = 11.84SRR5 pKa = 11.84NWCFTYY11 pKa = 10.93NNYY14 pKa = 10.46DD15 pKa = 3.28SSPRR19 pKa = 11.84IQKK22 pKa = 9.5CMKK25 pKa = 8.58YY26 pKa = 8.75LTYY29 pKa = 10.05GYY31 pKa = 10.22EE32 pKa = 4.13VGEE35 pKa = 4.38SGTPHH40 pKa = 6.23LQGFVIFKK48 pKa = 10.37NAVAKK53 pKa = 10.07PSQYY57 pKa = 10.52FKK59 pKa = 10.39PAYY62 pKa = 9.51HH63 pKa = 6.33FEE65 pKa = 4.33KK66 pKa = 10.88ARR68 pKa = 11.84GTPQQALEE76 pKa = 4.09YY77 pKa = 9.06CQKK80 pKa = 10.96DD81 pKa = 3.74GNFKK85 pKa = 10.9EE86 pKa = 5.01FGEE89 pKa = 4.41KK90 pKa = 10.06PKK92 pKa = 10.42TVADD96 pKa = 4.08GGAAGGEE103 pKa = 4.23ANKK106 pKa = 10.03RR107 pKa = 11.84RR108 pKa = 11.84YY109 pKa = 9.66EE110 pKa = 4.1EE111 pKa = 4.15AFSAAKK117 pKa = 9.47EE118 pKa = 3.99GRR120 pKa = 11.84MDD122 pKa = 6.65DD123 pKa = 3.7IPADD127 pKa = 3.17IYY129 pKa = 10.48IRR131 pKa = 11.84HH132 pKa = 6.06YY133 pKa = 10.24STLKK137 pKa = 10.52KK138 pKa = 10.1IRR140 pKa = 11.84FDD142 pKa = 3.67HH143 pKa = 7.29APPAQNNDD151 pKa = 2.93VLNNYY156 pKa = 7.25WVYY159 pKa = 11.03GPSGTGKK166 pKa = 9.65SKK168 pKa = 10.42SVRR171 pKa = 11.84EE172 pKa = 4.07FFGKK176 pKa = 9.85SLYY179 pKa = 10.56VKK181 pKa = 10.22NQNKK185 pKa = 8.09WFDD188 pKa = 3.68GYY190 pKa = 10.93EE191 pKa = 4.21GEE193 pKa = 5.11DD194 pKa = 3.53FVLIDD199 pKa = 4.53DD200 pKa = 4.5VHH202 pKa = 6.64PNWSGKK208 pKa = 8.88TILKK212 pKa = 9.42IWSDD216 pKa = 4.05HH217 pKa = 5.82YY218 pKa = 11.05PFSPEE223 pKa = 3.73TKK225 pKa = 9.5GGHH228 pKa = 5.51IKK230 pKa = 10.07MIRR233 pKa = 11.84PEE235 pKa = 4.47GIIVTSNYY243 pKa = 9.41TIEE246 pKa = 4.15EE247 pKa = 4.21MYY249 pKa = 10.47EE250 pKa = 4.0AEE252 pKa = 5.16EE253 pKa = 4.07DD254 pKa = 3.51RR255 pKa = 11.84QPIRR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84FKK263 pKa = 10.39VIKK266 pKa = 10.46SEE268 pKa = 3.89AGYY271 pKa = 10.7DD272 pKa = 3.83VIMKK276 pKa = 9.25EE277 pKa = 3.94YY278 pKa = 9.48NKK280 pKa = 10.19RR281 pKa = 11.84VAATKK286 pKa = 9.79ICSNTDD292 pKa = 2.73EE293 pKa = 4.6DD294 pKa = 4.71VVVAEE299 pKa = 5.19HH300 pKa = 6.42EE301 pKa = 4.23PCRR304 pKa = 11.84PATRR308 pKa = 11.84ASAPGDD314 pKa = 3.63HH315 pKa = 6.94EE316 pKa = 4.89LPPPATEE323 pKa = 3.89VQSIVVAVIEE333 pKa = 4.74SPPPPATTDD342 pKa = 3.12QQ343 pKa = 3.5
Molecular weight: 38.95 kDa
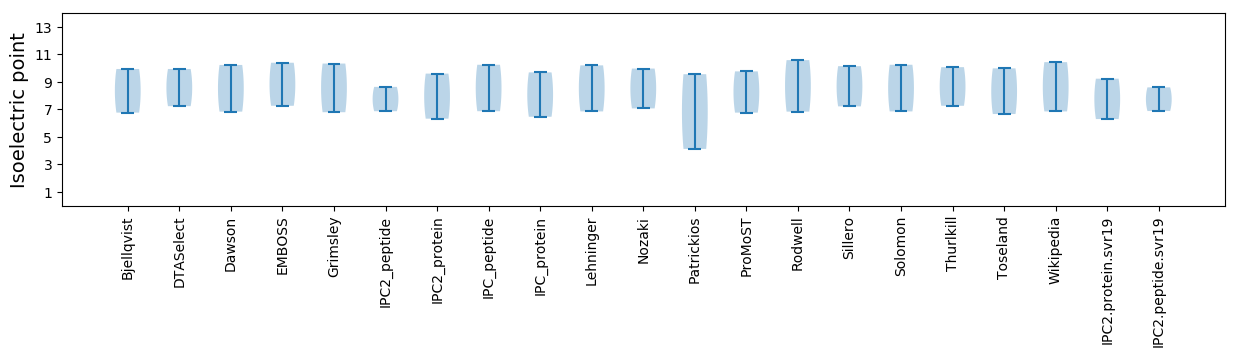
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6GII6|C6GII6_9VIRU Uncharacterized protein OS=Circovirus-like genome CB-A OX=642256 PE=4 SV=1
MM1 pKa = 6.65YY2 pKa = 10.05RR3 pKa = 11.84YY4 pKa = 9.18YY5 pKa = 10.86KK6 pKa = 10.25KK7 pKa = 10.62KK8 pKa = 9.18NAVRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84ATLKK18 pKa = 10.4RR19 pKa = 11.84SFKK22 pKa = 10.33RR23 pKa = 11.84GFRR26 pKa = 11.84RR27 pKa = 11.84AYY29 pKa = 8.79TPKK32 pKa = 10.29KK33 pKa = 9.78SYY35 pKa = 10.4RR36 pKa = 11.84PYY38 pKa = 10.99RR39 pKa = 11.84KK40 pKa = 9.32FKK42 pKa = 10.88SVNPKK47 pKa = 10.07IKK49 pKa = 10.4VPNIKK54 pKa = 10.22VPIAFSSTVALSTSADD70 pKa = 3.57KK71 pKa = 11.04IFTPEE76 pKa = 3.64PTLKK80 pKa = 10.87DD81 pKa = 3.61NEE83 pKa = 4.24VHH85 pKa = 6.26DD86 pKa = 4.43TYY88 pKa = 11.36TRR90 pKa = 11.84VYY92 pKa = 9.56NKK94 pKa = 9.88HH95 pKa = 5.71KK96 pKa = 10.53VVSIVNYY103 pKa = 7.5VQPHH107 pKa = 5.39QYY109 pKa = 10.52SSYY112 pKa = 10.26YY113 pKa = 9.55YY114 pKa = 9.42NLVGNGEE121 pKa = 4.05PLKK124 pKa = 10.46YY125 pKa = 10.04VVYY128 pKa = 10.67KK129 pKa = 9.61ITPDD133 pKa = 3.42TLGDD137 pKa = 3.51EE138 pKa = 4.61SKK140 pKa = 9.27PTNFRR145 pKa = 11.84EE146 pKa = 4.1AMAMPGARR154 pKa = 11.84AYY156 pKa = 11.18SLTRR160 pKa = 11.84SSKK163 pKa = 10.14RR164 pKa = 11.84MRR166 pKa = 11.84IAQMGVVNTYY176 pKa = 10.82LKK178 pKa = 10.86GSEE181 pKa = 4.15FVDD184 pKa = 3.59ANLTNVRR191 pKa = 11.84QSRR194 pKa = 11.84RR195 pKa = 11.84ATYY198 pKa = 10.18IPTNTLEE205 pKa = 5.46SDD207 pKa = 3.66LPWYY211 pKa = 10.47FGFRR215 pKa = 11.84IWFPQLTGGTLNLPPPSFEE234 pKa = 4.33VISVLNVSYY243 pKa = 11.11IMPTNASLTTIKK255 pKa = 10.75KK256 pKa = 10.35EE257 pKa = 4.72DD258 pKa = 3.58DD259 pKa = 3.7TLPDD263 pKa = 3.13NMFNLNCNSDD273 pKa = 4.23CIFF276 pKa = 4.13
MM1 pKa = 6.65YY2 pKa = 10.05RR3 pKa = 11.84YY4 pKa = 9.18YY5 pKa = 10.86KK6 pKa = 10.25KK7 pKa = 10.62KK8 pKa = 9.18NAVRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84ATLKK18 pKa = 10.4RR19 pKa = 11.84SFKK22 pKa = 10.33RR23 pKa = 11.84GFRR26 pKa = 11.84RR27 pKa = 11.84AYY29 pKa = 8.79TPKK32 pKa = 10.29KK33 pKa = 9.78SYY35 pKa = 10.4RR36 pKa = 11.84PYY38 pKa = 10.99RR39 pKa = 11.84KK40 pKa = 9.32FKK42 pKa = 10.88SVNPKK47 pKa = 10.07IKK49 pKa = 10.4VPNIKK54 pKa = 10.22VPIAFSSTVALSTSADD70 pKa = 3.57KK71 pKa = 11.04IFTPEE76 pKa = 3.64PTLKK80 pKa = 10.87DD81 pKa = 3.61NEE83 pKa = 4.24VHH85 pKa = 6.26DD86 pKa = 4.43TYY88 pKa = 11.36TRR90 pKa = 11.84VYY92 pKa = 9.56NKK94 pKa = 9.88HH95 pKa = 5.71KK96 pKa = 10.53VVSIVNYY103 pKa = 7.5VQPHH107 pKa = 5.39QYY109 pKa = 10.52SSYY112 pKa = 10.26YY113 pKa = 9.55YY114 pKa = 9.42NLVGNGEE121 pKa = 4.05PLKK124 pKa = 10.46YY125 pKa = 10.04VVYY128 pKa = 10.67KK129 pKa = 9.61ITPDD133 pKa = 3.42TLGDD137 pKa = 3.51EE138 pKa = 4.61SKK140 pKa = 9.27PTNFRR145 pKa = 11.84EE146 pKa = 4.1AMAMPGARR154 pKa = 11.84AYY156 pKa = 11.18SLTRR160 pKa = 11.84SSKK163 pKa = 10.14RR164 pKa = 11.84MRR166 pKa = 11.84IAQMGVVNTYY176 pKa = 10.82LKK178 pKa = 10.86GSEE181 pKa = 4.15FVDD184 pKa = 3.59ANLTNVRR191 pKa = 11.84QSRR194 pKa = 11.84RR195 pKa = 11.84ATYY198 pKa = 10.18IPTNTLEE205 pKa = 5.46SDD207 pKa = 3.66LPWYY211 pKa = 10.47FGFRR215 pKa = 11.84IWFPQLTGGTLNLPPPSFEE234 pKa = 4.33VISVLNVSYY243 pKa = 11.11IMPTNASLTTIKK255 pKa = 10.75KK256 pKa = 10.35EE257 pKa = 4.72DD258 pKa = 3.58DD259 pKa = 3.7TLPDD263 pKa = 3.13NMFNLNCNSDD273 pKa = 4.23CIFF276 pKa = 4.13
Molecular weight: 31.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
619 |
276 |
343 |
309.5 |
35.41 |
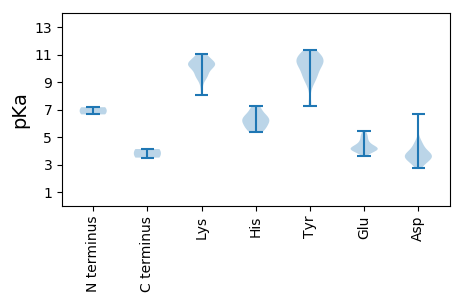
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.462 ± 0.853 | 1.131 ± 0.249 |
4.847 ± 0.529 | 5.977 ± 1.668 |
4.523 ± 0.115 | 5.493 ± 1.148 |
1.939 ± 0.523 | 5.331 ± 0.381 |
8.239 ± 0.058 | 4.362 ± 1.326 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.1 ± 0.268 | 5.816 ± 0.878 |
7.27 ± 0.014 | 2.585 ± 0.475 |
6.139 ± 0.68 | 6.947 ± 0.629 |
6.462 ± 1.149 | 6.785 ± 0.506 |
1.131 ± 0.249 | 6.462 ± 0.482 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
