
Hubei narna-like virus 24
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.32
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIB4|A0A1L3KIB4_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 24 OX=1922955 PE=4 SV=1
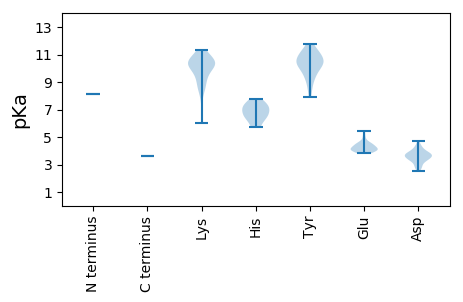
MM1 pKa = 8.13DD2 pKa = 3.55NTITHH7 pKa = 7.21ALCYY11 pKa = 9.64AANITLGKK19 pKa = 9.63DD20 pKa = 2.94AKK22 pKa = 10.55PFIEE26 pKa = 5.11RR27 pKa = 11.84IEE29 pKa = 4.45HH30 pKa = 5.7FLSHH34 pKa = 6.87NGTEE38 pKa = 4.24YY39 pKa = 10.37TVNRR43 pKa = 11.84LKK45 pKa = 10.9VIYY48 pKa = 10.05QGALLLRR55 pKa = 11.84EE56 pKa = 4.12YY57 pKa = 10.51DD58 pKa = 3.47QLGRR62 pKa = 11.84PVDD65 pKa = 3.64TDD67 pKa = 3.64FLGNTNRR74 pKa = 11.84GSFDD78 pKa = 3.42QAVKK82 pKa = 10.4LYY84 pKa = 9.92HH85 pKa = 6.63EE86 pKa = 4.85NGIATSKK93 pKa = 10.46KK94 pKa = 10.25GYY96 pKa = 9.24PKK98 pKa = 10.51DD99 pKa = 3.29STGIVVRR106 pKa = 11.84QFVEE110 pKa = 4.25SNRR113 pKa = 11.84PQVIRR118 pKa = 11.84NMTGLLRR125 pKa = 11.84SYY127 pKa = 9.11TQFRR131 pKa = 11.84LDD133 pKa = 3.75SVSQQQVDD141 pKa = 3.74KK142 pKa = 10.87VVEE145 pKa = 4.56SVCMPRR151 pKa = 11.84DD152 pKa = 3.32RR153 pKa = 11.84AHH155 pKa = 7.77DD156 pKa = 3.87EE157 pKa = 3.92NQLFDD162 pKa = 4.22NLIHH166 pKa = 7.35LSRR169 pKa = 11.84VPKK172 pKa = 10.51ALWNKK177 pKa = 9.46SIKK180 pKa = 10.37AEE182 pKa = 4.04RR183 pKa = 11.84LSMDD187 pKa = 3.78CLRR190 pKa = 11.84PYY192 pKa = 10.09TSVYY196 pKa = 10.41CHH198 pKa = 6.54GSVPKK203 pKa = 10.15HH204 pKa = 6.25VKK206 pKa = 10.29DD207 pKa = 3.73EE208 pKa = 4.6PYY210 pKa = 11.12GKK212 pKa = 10.11ALYY215 pKa = 11.24SMITTSYY222 pKa = 10.37LPSPLEE228 pKa = 3.81SVVPSKK234 pKa = 10.77SIRR237 pKa = 11.84DD238 pKa = 3.73RR239 pKa = 11.84VRR241 pKa = 11.84SWMPRR246 pKa = 11.84EE247 pKa = 3.86NDD249 pKa = 3.38YY250 pKa = 11.59DD251 pKa = 3.76PEE253 pKa = 4.1QYY255 pKa = 10.42AGKK258 pKa = 9.5IHH260 pKa = 7.37IIQEE264 pKa = 4.07AGAKK268 pKa = 9.44ARR270 pKa = 11.84TVAIPNGWTQIGFVPLHH287 pKa = 6.78DD288 pKa = 3.84QMQLEE293 pKa = 4.47SEE295 pKa = 4.56RR296 pKa = 11.84YY297 pKa = 8.48FPSEE301 pKa = 3.82SCVFDD306 pKa = 3.59QLKK309 pKa = 10.67GIYY312 pKa = 10.0ALRR315 pKa = 11.84NHH317 pKa = 6.4MGDD320 pKa = 2.55GKK322 pKa = 9.21TPVSVDD328 pKa = 3.47LSSATDD334 pKa = 3.6RR335 pKa = 11.84FPIEE339 pKa = 4.25FQCKK343 pKa = 8.47LLSHH347 pKa = 7.12IGLPFYY353 pKa = 10.94SDD355 pKa = 3.51SLSEE359 pKa = 4.05LVKK362 pKa = 10.9KK363 pKa = 10.07PWEE366 pKa = 4.31FPQGKK371 pKa = 8.37PFGYY375 pKa = 10.66DD376 pKa = 2.66SVYY379 pKa = 11.09YY380 pKa = 10.54NAGQPMGLYY389 pKa = 10.38GSFPTFHH396 pKa = 7.21FSNLVMADD404 pKa = 3.75ASCRR408 pKa = 11.84LTDD411 pKa = 3.64YY412 pKa = 11.18QIGEE416 pKa = 4.43TIDD419 pKa = 3.83GKK421 pKa = 11.2GKK423 pKa = 9.31EE424 pKa = 4.16QVAGIVYY431 pKa = 8.65EE432 pKa = 4.03RR433 pKa = 11.84LYY435 pKa = 11.24GFYY438 pKa = 10.75AHH440 pKa = 7.66RR441 pKa = 11.84DD442 pKa = 3.32KK443 pKa = 11.34DD444 pKa = 4.33TVPDD448 pKa = 4.73DD449 pKa = 4.04YY450 pKa = 11.45IEE452 pKa = 4.47RR453 pKa = 11.84NNILGNAAGDD463 pKa = 4.01SPVSKK468 pKa = 10.26FYY470 pKa = 10.97DD471 pKa = 3.5GSRR474 pKa = 11.84FKK476 pKa = 11.36VLGDD480 pKa = 4.48DD481 pKa = 3.96IVFSDD486 pKa = 4.05EE487 pKa = 4.18RR488 pKa = 11.84DD489 pKa = 3.24AAHH492 pKa = 5.73YY493 pKa = 7.92TKK495 pKa = 10.39IMKK498 pKa = 8.63TINVPISHH506 pKa = 6.87HH507 pKa = 6.26KK508 pKa = 10.31SFSKK512 pKa = 10.9GMVCEE517 pKa = 4.15FAGAIAMRR525 pKa = 11.84TKK527 pKa = 10.62KK528 pKa = 10.46SNKK531 pKa = 8.4DD532 pKa = 2.88RR533 pKa = 11.84RR534 pKa = 11.84YY535 pKa = 9.77DD536 pKa = 3.37VSIFRR541 pKa = 11.84PYY543 pKa = 10.76KK544 pKa = 9.47FPKK547 pKa = 10.14DD548 pKa = 3.26GSFIGNPISFIYY560 pKa = 10.35AFSSPQSKK568 pKa = 9.76QRR570 pKa = 11.84VSKK573 pKa = 9.95KK574 pKa = 7.69WKK576 pKa = 9.99QFFEE580 pKa = 5.47DD581 pKa = 3.79FDD583 pKa = 3.53KK584 pKa = 11.1TRR586 pKa = 11.84PQRR589 pKa = 11.84YY590 pKa = 9.4ADD592 pKa = 4.31LCPITPFVEE601 pKa = 4.0DD602 pKa = 2.75RR603 pKa = 11.84VGIVGRR609 pKa = 11.84FRR611 pKa = 11.84DD612 pKa = 3.54QDD614 pKa = 2.83IDD616 pKa = 3.78RR617 pKa = 11.84VVDD620 pKa = 3.99NIYY623 pKa = 10.63RR624 pKa = 11.84FSNLSFFPYY633 pKa = 10.19TDD635 pKa = 3.22TGKK638 pKa = 8.81DD639 pKa = 3.13TPIIYY644 pKa = 9.87DD645 pKa = 3.7GPRR648 pKa = 11.84NRR650 pKa = 11.84APLYY654 pKa = 9.31QVPLFIKK661 pKa = 10.26KK662 pKa = 10.13KK663 pKa = 8.82GIEE666 pKa = 4.03VPGFEE671 pKa = 4.43TQRR674 pKa = 11.84PASDD678 pKa = 2.92TRR680 pKa = 11.84SYY682 pKa = 11.78AEE684 pKa = 4.01TVKK687 pKa = 10.66RR688 pKa = 11.84LYY690 pKa = 10.75SDD692 pKa = 4.2PLMKK696 pKa = 10.43AQQKK700 pKa = 6.03TRR702 pKa = 11.84KK703 pKa = 8.91SSAMKK708 pKa = 10.13II709 pKa = 3.65
MM1 pKa = 8.13DD2 pKa = 3.55NTITHH7 pKa = 7.21ALCYY11 pKa = 9.64AANITLGKK19 pKa = 9.63DD20 pKa = 2.94AKK22 pKa = 10.55PFIEE26 pKa = 5.11RR27 pKa = 11.84IEE29 pKa = 4.45HH30 pKa = 5.7FLSHH34 pKa = 6.87NGTEE38 pKa = 4.24YY39 pKa = 10.37TVNRR43 pKa = 11.84LKK45 pKa = 10.9VIYY48 pKa = 10.05QGALLLRR55 pKa = 11.84EE56 pKa = 4.12YY57 pKa = 10.51DD58 pKa = 3.47QLGRR62 pKa = 11.84PVDD65 pKa = 3.64TDD67 pKa = 3.64FLGNTNRR74 pKa = 11.84GSFDD78 pKa = 3.42QAVKK82 pKa = 10.4LYY84 pKa = 9.92HH85 pKa = 6.63EE86 pKa = 4.85NGIATSKK93 pKa = 10.46KK94 pKa = 10.25GYY96 pKa = 9.24PKK98 pKa = 10.51DD99 pKa = 3.29STGIVVRR106 pKa = 11.84QFVEE110 pKa = 4.25SNRR113 pKa = 11.84PQVIRR118 pKa = 11.84NMTGLLRR125 pKa = 11.84SYY127 pKa = 9.11TQFRR131 pKa = 11.84LDD133 pKa = 3.75SVSQQQVDD141 pKa = 3.74KK142 pKa = 10.87VVEE145 pKa = 4.56SVCMPRR151 pKa = 11.84DD152 pKa = 3.32RR153 pKa = 11.84AHH155 pKa = 7.77DD156 pKa = 3.87EE157 pKa = 3.92NQLFDD162 pKa = 4.22NLIHH166 pKa = 7.35LSRR169 pKa = 11.84VPKK172 pKa = 10.51ALWNKK177 pKa = 9.46SIKK180 pKa = 10.37AEE182 pKa = 4.04RR183 pKa = 11.84LSMDD187 pKa = 3.78CLRR190 pKa = 11.84PYY192 pKa = 10.09TSVYY196 pKa = 10.41CHH198 pKa = 6.54GSVPKK203 pKa = 10.15HH204 pKa = 6.25VKK206 pKa = 10.29DD207 pKa = 3.73EE208 pKa = 4.6PYY210 pKa = 11.12GKK212 pKa = 10.11ALYY215 pKa = 11.24SMITTSYY222 pKa = 10.37LPSPLEE228 pKa = 3.81SVVPSKK234 pKa = 10.77SIRR237 pKa = 11.84DD238 pKa = 3.73RR239 pKa = 11.84VRR241 pKa = 11.84SWMPRR246 pKa = 11.84EE247 pKa = 3.86NDD249 pKa = 3.38YY250 pKa = 11.59DD251 pKa = 3.76PEE253 pKa = 4.1QYY255 pKa = 10.42AGKK258 pKa = 9.5IHH260 pKa = 7.37IIQEE264 pKa = 4.07AGAKK268 pKa = 9.44ARR270 pKa = 11.84TVAIPNGWTQIGFVPLHH287 pKa = 6.78DD288 pKa = 3.84QMQLEE293 pKa = 4.47SEE295 pKa = 4.56RR296 pKa = 11.84YY297 pKa = 8.48FPSEE301 pKa = 3.82SCVFDD306 pKa = 3.59QLKK309 pKa = 10.67GIYY312 pKa = 10.0ALRR315 pKa = 11.84NHH317 pKa = 6.4MGDD320 pKa = 2.55GKK322 pKa = 9.21TPVSVDD328 pKa = 3.47LSSATDD334 pKa = 3.6RR335 pKa = 11.84FPIEE339 pKa = 4.25FQCKK343 pKa = 8.47LLSHH347 pKa = 7.12IGLPFYY353 pKa = 10.94SDD355 pKa = 3.51SLSEE359 pKa = 4.05LVKK362 pKa = 10.9KK363 pKa = 10.07PWEE366 pKa = 4.31FPQGKK371 pKa = 8.37PFGYY375 pKa = 10.66DD376 pKa = 2.66SVYY379 pKa = 11.09YY380 pKa = 10.54NAGQPMGLYY389 pKa = 10.38GSFPTFHH396 pKa = 7.21FSNLVMADD404 pKa = 3.75ASCRR408 pKa = 11.84LTDD411 pKa = 3.64YY412 pKa = 11.18QIGEE416 pKa = 4.43TIDD419 pKa = 3.83GKK421 pKa = 11.2GKK423 pKa = 9.31EE424 pKa = 4.16QVAGIVYY431 pKa = 8.65EE432 pKa = 4.03RR433 pKa = 11.84LYY435 pKa = 11.24GFYY438 pKa = 10.75AHH440 pKa = 7.66RR441 pKa = 11.84DD442 pKa = 3.32KK443 pKa = 11.34DD444 pKa = 4.33TVPDD448 pKa = 4.73DD449 pKa = 4.04YY450 pKa = 11.45IEE452 pKa = 4.47RR453 pKa = 11.84NNILGNAAGDD463 pKa = 4.01SPVSKK468 pKa = 10.26FYY470 pKa = 10.97DD471 pKa = 3.5GSRR474 pKa = 11.84FKK476 pKa = 11.36VLGDD480 pKa = 4.48DD481 pKa = 3.96IVFSDD486 pKa = 4.05EE487 pKa = 4.18RR488 pKa = 11.84DD489 pKa = 3.24AAHH492 pKa = 5.73YY493 pKa = 7.92TKK495 pKa = 10.39IMKK498 pKa = 8.63TINVPISHH506 pKa = 6.87HH507 pKa = 6.26KK508 pKa = 10.31SFSKK512 pKa = 10.9GMVCEE517 pKa = 4.15FAGAIAMRR525 pKa = 11.84TKK527 pKa = 10.62KK528 pKa = 10.46SNKK531 pKa = 8.4DD532 pKa = 2.88RR533 pKa = 11.84RR534 pKa = 11.84YY535 pKa = 9.77DD536 pKa = 3.37VSIFRR541 pKa = 11.84PYY543 pKa = 10.76KK544 pKa = 9.47FPKK547 pKa = 10.14DD548 pKa = 3.26GSFIGNPISFIYY560 pKa = 10.35AFSSPQSKK568 pKa = 9.76QRR570 pKa = 11.84VSKK573 pKa = 9.95KK574 pKa = 7.69WKK576 pKa = 9.99QFFEE580 pKa = 5.47DD581 pKa = 3.79FDD583 pKa = 3.53KK584 pKa = 11.1TRR586 pKa = 11.84PQRR589 pKa = 11.84YY590 pKa = 9.4ADD592 pKa = 4.31LCPITPFVEE601 pKa = 4.0DD602 pKa = 2.75RR603 pKa = 11.84VGIVGRR609 pKa = 11.84FRR611 pKa = 11.84DD612 pKa = 3.54QDD614 pKa = 2.83IDD616 pKa = 3.78RR617 pKa = 11.84VVDD620 pKa = 3.99NIYY623 pKa = 10.63RR624 pKa = 11.84FSNLSFFPYY633 pKa = 10.19TDD635 pKa = 3.22TGKK638 pKa = 8.81DD639 pKa = 3.13TPIIYY644 pKa = 9.87DD645 pKa = 3.7GPRR648 pKa = 11.84NRR650 pKa = 11.84APLYY654 pKa = 9.31QVPLFIKK661 pKa = 10.26KK662 pKa = 10.13KK663 pKa = 8.82GIEE666 pKa = 4.03VPGFEE671 pKa = 4.43TQRR674 pKa = 11.84PASDD678 pKa = 2.92TRR680 pKa = 11.84SYY682 pKa = 11.78AEE684 pKa = 4.01TVKK687 pKa = 10.66RR688 pKa = 11.84LYY690 pKa = 10.75SDD692 pKa = 4.2PLMKK696 pKa = 10.43AQQKK700 pKa = 6.03TRR702 pKa = 11.84KK703 pKa = 8.91SSAMKK708 pKa = 10.13II709 pKa = 3.65
Molecular weight: 81.03 kDa
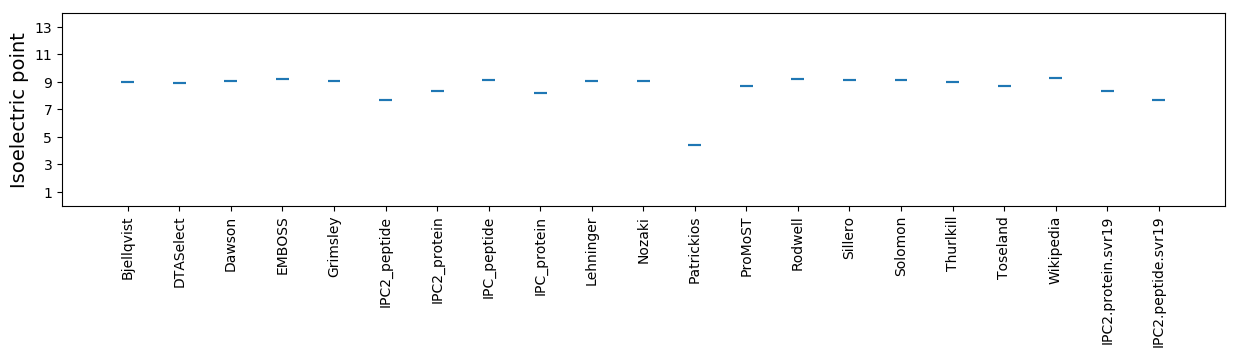
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIB4|A0A1L3KIB4_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 24 OX=1922955 PE=4 SV=1
MM1 pKa = 8.13DD2 pKa = 3.55NTITHH7 pKa = 7.21ALCYY11 pKa = 9.64AANITLGKK19 pKa = 9.63DD20 pKa = 2.94AKK22 pKa = 10.55PFIEE26 pKa = 5.11RR27 pKa = 11.84IEE29 pKa = 4.45HH30 pKa = 5.7FLSHH34 pKa = 6.87NGTEE38 pKa = 4.24YY39 pKa = 10.37TVNRR43 pKa = 11.84LKK45 pKa = 10.9VIYY48 pKa = 10.05QGALLLRR55 pKa = 11.84EE56 pKa = 4.12YY57 pKa = 10.51DD58 pKa = 3.47QLGRR62 pKa = 11.84PVDD65 pKa = 3.64TDD67 pKa = 3.64FLGNTNRR74 pKa = 11.84GSFDD78 pKa = 3.42QAVKK82 pKa = 10.4LYY84 pKa = 9.92HH85 pKa = 6.63EE86 pKa = 4.85NGIATSKK93 pKa = 10.46KK94 pKa = 10.25GYY96 pKa = 9.24PKK98 pKa = 10.51DD99 pKa = 3.29STGIVVRR106 pKa = 11.84QFVEE110 pKa = 4.25SNRR113 pKa = 11.84PQVIRR118 pKa = 11.84NMTGLLRR125 pKa = 11.84SYY127 pKa = 9.11TQFRR131 pKa = 11.84LDD133 pKa = 3.75SVSQQQVDD141 pKa = 3.74KK142 pKa = 10.87VVEE145 pKa = 4.56SVCMPRR151 pKa = 11.84DD152 pKa = 3.32RR153 pKa = 11.84AHH155 pKa = 7.77DD156 pKa = 3.87EE157 pKa = 3.92NQLFDD162 pKa = 4.22NLIHH166 pKa = 7.35LSRR169 pKa = 11.84VPKK172 pKa = 10.51ALWNKK177 pKa = 9.46SIKK180 pKa = 10.37AEE182 pKa = 4.04RR183 pKa = 11.84LSMDD187 pKa = 3.78CLRR190 pKa = 11.84PYY192 pKa = 10.09TSVYY196 pKa = 10.41CHH198 pKa = 6.54GSVPKK203 pKa = 10.15HH204 pKa = 6.25VKK206 pKa = 10.29DD207 pKa = 3.73EE208 pKa = 4.6PYY210 pKa = 11.12GKK212 pKa = 10.11ALYY215 pKa = 11.24SMITTSYY222 pKa = 10.37LPSPLEE228 pKa = 3.81SVVPSKK234 pKa = 10.77SIRR237 pKa = 11.84DD238 pKa = 3.73RR239 pKa = 11.84VRR241 pKa = 11.84SWMPRR246 pKa = 11.84EE247 pKa = 3.86NDD249 pKa = 3.38YY250 pKa = 11.59DD251 pKa = 3.76PEE253 pKa = 4.1QYY255 pKa = 10.42AGKK258 pKa = 9.5IHH260 pKa = 7.37IIQEE264 pKa = 4.07AGAKK268 pKa = 9.44ARR270 pKa = 11.84TVAIPNGWTQIGFVPLHH287 pKa = 6.78DD288 pKa = 3.84QMQLEE293 pKa = 4.47SEE295 pKa = 4.56RR296 pKa = 11.84YY297 pKa = 8.48FPSEE301 pKa = 3.82SCVFDD306 pKa = 3.59QLKK309 pKa = 10.67GIYY312 pKa = 10.0ALRR315 pKa = 11.84NHH317 pKa = 6.4MGDD320 pKa = 2.55GKK322 pKa = 9.21TPVSVDD328 pKa = 3.47LSSATDD334 pKa = 3.6RR335 pKa = 11.84FPIEE339 pKa = 4.25FQCKK343 pKa = 8.47LLSHH347 pKa = 7.12IGLPFYY353 pKa = 10.94SDD355 pKa = 3.51SLSEE359 pKa = 4.05LVKK362 pKa = 10.9KK363 pKa = 10.07PWEE366 pKa = 4.31FPQGKK371 pKa = 8.37PFGYY375 pKa = 10.66DD376 pKa = 2.66SVYY379 pKa = 11.09YY380 pKa = 10.54NAGQPMGLYY389 pKa = 10.38GSFPTFHH396 pKa = 7.21FSNLVMADD404 pKa = 3.75ASCRR408 pKa = 11.84LTDD411 pKa = 3.64YY412 pKa = 11.18QIGEE416 pKa = 4.43TIDD419 pKa = 3.83GKK421 pKa = 11.2GKK423 pKa = 9.31EE424 pKa = 4.16QVAGIVYY431 pKa = 8.65EE432 pKa = 4.03RR433 pKa = 11.84LYY435 pKa = 11.24GFYY438 pKa = 10.75AHH440 pKa = 7.66RR441 pKa = 11.84DD442 pKa = 3.32KK443 pKa = 11.34DD444 pKa = 4.33TVPDD448 pKa = 4.73DD449 pKa = 4.04YY450 pKa = 11.45IEE452 pKa = 4.47RR453 pKa = 11.84NNILGNAAGDD463 pKa = 4.01SPVSKK468 pKa = 10.26FYY470 pKa = 10.97DD471 pKa = 3.5GSRR474 pKa = 11.84FKK476 pKa = 11.36VLGDD480 pKa = 4.48DD481 pKa = 3.96IVFSDD486 pKa = 4.05EE487 pKa = 4.18RR488 pKa = 11.84DD489 pKa = 3.24AAHH492 pKa = 5.73YY493 pKa = 7.92TKK495 pKa = 10.39IMKK498 pKa = 8.63TINVPISHH506 pKa = 6.87HH507 pKa = 6.26KK508 pKa = 10.31SFSKK512 pKa = 10.9GMVCEE517 pKa = 4.15FAGAIAMRR525 pKa = 11.84TKK527 pKa = 10.62KK528 pKa = 10.46SNKK531 pKa = 8.4DD532 pKa = 2.88RR533 pKa = 11.84RR534 pKa = 11.84YY535 pKa = 9.77DD536 pKa = 3.37VSIFRR541 pKa = 11.84PYY543 pKa = 10.76KK544 pKa = 9.47FPKK547 pKa = 10.14DD548 pKa = 3.26GSFIGNPISFIYY560 pKa = 10.35AFSSPQSKK568 pKa = 9.76QRR570 pKa = 11.84VSKK573 pKa = 9.95KK574 pKa = 7.69WKK576 pKa = 9.99QFFEE580 pKa = 5.47DD581 pKa = 3.79FDD583 pKa = 3.53KK584 pKa = 11.1TRR586 pKa = 11.84PQRR589 pKa = 11.84YY590 pKa = 9.4ADD592 pKa = 4.31LCPITPFVEE601 pKa = 4.0DD602 pKa = 2.75RR603 pKa = 11.84VGIVGRR609 pKa = 11.84FRR611 pKa = 11.84DD612 pKa = 3.54QDD614 pKa = 2.83IDD616 pKa = 3.78RR617 pKa = 11.84VVDD620 pKa = 3.99NIYY623 pKa = 10.63RR624 pKa = 11.84FSNLSFFPYY633 pKa = 10.19TDD635 pKa = 3.22TGKK638 pKa = 8.81DD639 pKa = 3.13TPIIYY644 pKa = 9.87DD645 pKa = 3.7GPRR648 pKa = 11.84NRR650 pKa = 11.84APLYY654 pKa = 9.31QVPLFIKK661 pKa = 10.26KK662 pKa = 10.13KK663 pKa = 8.82GIEE666 pKa = 4.03VPGFEE671 pKa = 4.43TQRR674 pKa = 11.84PASDD678 pKa = 2.92TRR680 pKa = 11.84SYY682 pKa = 11.78AEE684 pKa = 4.01TVKK687 pKa = 10.66RR688 pKa = 11.84LYY690 pKa = 10.75SDD692 pKa = 4.2PLMKK696 pKa = 10.43AQQKK700 pKa = 6.03TRR702 pKa = 11.84KK703 pKa = 8.91SSAMKK708 pKa = 10.13II709 pKa = 3.65
MM1 pKa = 8.13DD2 pKa = 3.55NTITHH7 pKa = 7.21ALCYY11 pKa = 9.64AANITLGKK19 pKa = 9.63DD20 pKa = 2.94AKK22 pKa = 10.55PFIEE26 pKa = 5.11RR27 pKa = 11.84IEE29 pKa = 4.45HH30 pKa = 5.7FLSHH34 pKa = 6.87NGTEE38 pKa = 4.24YY39 pKa = 10.37TVNRR43 pKa = 11.84LKK45 pKa = 10.9VIYY48 pKa = 10.05QGALLLRR55 pKa = 11.84EE56 pKa = 4.12YY57 pKa = 10.51DD58 pKa = 3.47QLGRR62 pKa = 11.84PVDD65 pKa = 3.64TDD67 pKa = 3.64FLGNTNRR74 pKa = 11.84GSFDD78 pKa = 3.42QAVKK82 pKa = 10.4LYY84 pKa = 9.92HH85 pKa = 6.63EE86 pKa = 4.85NGIATSKK93 pKa = 10.46KK94 pKa = 10.25GYY96 pKa = 9.24PKK98 pKa = 10.51DD99 pKa = 3.29STGIVVRR106 pKa = 11.84QFVEE110 pKa = 4.25SNRR113 pKa = 11.84PQVIRR118 pKa = 11.84NMTGLLRR125 pKa = 11.84SYY127 pKa = 9.11TQFRR131 pKa = 11.84LDD133 pKa = 3.75SVSQQQVDD141 pKa = 3.74KK142 pKa = 10.87VVEE145 pKa = 4.56SVCMPRR151 pKa = 11.84DD152 pKa = 3.32RR153 pKa = 11.84AHH155 pKa = 7.77DD156 pKa = 3.87EE157 pKa = 3.92NQLFDD162 pKa = 4.22NLIHH166 pKa = 7.35LSRR169 pKa = 11.84VPKK172 pKa = 10.51ALWNKK177 pKa = 9.46SIKK180 pKa = 10.37AEE182 pKa = 4.04RR183 pKa = 11.84LSMDD187 pKa = 3.78CLRR190 pKa = 11.84PYY192 pKa = 10.09TSVYY196 pKa = 10.41CHH198 pKa = 6.54GSVPKK203 pKa = 10.15HH204 pKa = 6.25VKK206 pKa = 10.29DD207 pKa = 3.73EE208 pKa = 4.6PYY210 pKa = 11.12GKK212 pKa = 10.11ALYY215 pKa = 11.24SMITTSYY222 pKa = 10.37LPSPLEE228 pKa = 3.81SVVPSKK234 pKa = 10.77SIRR237 pKa = 11.84DD238 pKa = 3.73RR239 pKa = 11.84VRR241 pKa = 11.84SWMPRR246 pKa = 11.84EE247 pKa = 3.86NDD249 pKa = 3.38YY250 pKa = 11.59DD251 pKa = 3.76PEE253 pKa = 4.1QYY255 pKa = 10.42AGKK258 pKa = 9.5IHH260 pKa = 7.37IIQEE264 pKa = 4.07AGAKK268 pKa = 9.44ARR270 pKa = 11.84TVAIPNGWTQIGFVPLHH287 pKa = 6.78DD288 pKa = 3.84QMQLEE293 pKa = 4.47SEE295 pKa = 4.56RR296 pKa = 11.84YY297 pKa = 8.48FPSEE301 pKa = 3.82SCVFDD306 pKa = 3.59QLKK309 pKa = 10.67GIYY312 pKa = 10.0ALRR315 pKa = 11.84NHH317 pKa = 6.4MGDD320 pKa = 2.55GKK322 pKa = 9.21TPVSVDD328 pKa = 3.47LSSATDD334 pKa = 3.6RR335 pKa = 11.84FPIEE339 pKa = 4.25FQCKK343 pKa = 8.47LLSHH347 pKa = 7.12IGLPFYY353 pKa = 10.94SDD355 pKa = 3.51SLSEE359 pKa = 4.05LVKK362 pKa = 10.9KK363 pKa = 10.07PWEE366 pKa = 4.31FPQGKK371 pKa = 8.37PFGYY375 pKa = 10.66DD376 pKa = 2.66SVYY379 pKa = 11.09YY380 pKa = 10.54NAGQPMGLYY389 pKa = 10.38GSFPTFHH396 pKa = 7.21FSNLVMADD404 pKa = 3.75ASCRR408 pKa = 11.84LTDD411 pKa = 3.64YY412 pKa = 11.18QIGEE416 pKa = 4.43TIDD419 pKa = 3.83GKK421 pKa = 11.2GKK423 pKa = 9.31EE424 pKa = 4.16QVAGIVYY431 pKa = 8.65EE432 pKa = 4.03RR433 pKa = 11.84LYY435 pKa = 11.24GFYY438 pKa = 10.75AHH440 pKa = 7.66RR441 pKa = 11.84DD442 pKa = 3.32KK443 pKa = 11.34DD444 pKa = 4.33TVPDD448 pKa = 4.73DD449 pKa = 4.04YY450 pKa = 11.45IEE452 pKa = 4.47RR453 pKa = 11.84NNILGNAAGDD463 pKa = 4.01SPVSKK468 pKa = 10.26FYY470 pKa = 10.97DD471 pKa = 3.5GSRR474 pKa = 11.84FKK476 pKa = 11.36VLGDD480 pKa = 4.48DD481 pKa = 3.96IVFSDD486 pKa = 4.05EE487 pKa = 4.18RR488 pKa = 11.84DD489 pKa = 3.24AAHH492 pKa = 5.73YY493 pKa = 7.92TKK495 pKa = 10.39IMKK498 pKa = 8.63TINVPISHH506 pKa = 6.87HH507 pKa = 6.26KK508 pKa = 10.31SFSKK512 pKa = 10.9GMVCEE517 pKa = 4.15FAGAIAMRR525 pKa = 11.84TKK527 pKa = 10.62KK528 pKa = 10.46SNKK531 pKa = 8.4DD532 pKa = 2.88RR533 pKa = 11.84RR534 pKa = 11.84YY535 pKa = 9.77DD536 pKa = 3.37VSIFRR541 pKa = 11.84PYY543 pKa = 10.76KK544 pKa = 9.47FPKK547 pKa = 10.14DD548 pKa = 3.26GSFIGNPISFIYY560 pKa = 10.35AFSSPQSKK568 pKa = 9.76QRR570 pKa = 11.84VSKK573 pKa = 9.95KK574 pKa = 7.69WKK576 pKa = 9.99QFFEE580 pKa = 5.47DD581 pKa = 3.79FDD583 pKa = 3.53KK584 pKa = 11.1TRR586 pKa = 11.84PQRR589 pKa = 11.84YY590 pKa = 9.4ADD592 pKa = 4.31LCPITPFVEE601 pKa = 4.0DD602 pKa = 2.75RR603 pKa = 11.84VGIVGRR609 pKa = 11.84FRR611 pKa = 11.84DD612 pKa = 3.54QDD614 pKa = 2.83IDD616 pKa = 3.78RR617 pKa = 11.84VVDD620 pKa = 3.99NIYY623 pKa = 10.63RR624 pKa = 11.84FSNLSFFPYY633 pKa = 10.19TDD635 pKa = 3.22TGKK638 pKa = 8.81DD639 pKa = 3.13TPIIYY644 pKa = 9.87DD645 pKa = 3.7GPRR648 pKa = 11.84NRR650 pKa = 11.84APLYY654 pKa = 9.31QVPLFIKK661 pKa = 10.26KK662 pKa = 10.13KK663 pKa = 8.82GIEE666 pKa = 4.03VPGFEE671 pKa = 4.43TQRR674 pKa = 11.84PASDD678 pKa = 2.92TRR680 pKa = 11.84SYY682 pKa = 11.78AEE684 pKa = 4.01TVKK687 pKa = 10.66RR688 pKa = 11.84LYY690 pKa = 10.75SDD692 pKa = 4.2PLMKK696 pKa = 10.43AQQKK700 pKa = 6.03TRR702 pKa = 11.84KK703 pKa = 8.91SSAMKK708 pKa = 10.13II709 pKa = 3.65
Molecular weight: 81.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
709 |
709 |
709 |
709.0 |
81.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.219 ± 0.0 | 1.269 ± 0.0 |
7.475 ± 0.0 | 4.372 ± 0.0 |
5.501 ± 0.0 | 6.206 ± 0.0 |
2.398 ± 0.0 | 6.065 ± 0.0 |
7.193 ± 0.0 | 6.206 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.116 ± 0.0 | 3.667 ± 0.0 |
6.065 ± 0.0 | 4.231 ± 0.0 |
6.629 ± 0.0 | 8.039 ± 0.0 |
4.795 ± 0.0 | 6.488 ± 0.0 |
0.705 ± 0.0 | 5.36 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
