
Staphylococcus phage SA537ruMSSAST97
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Bronfenbrennervirinae; Peeveelvirus; unclassified Peeveelvirus
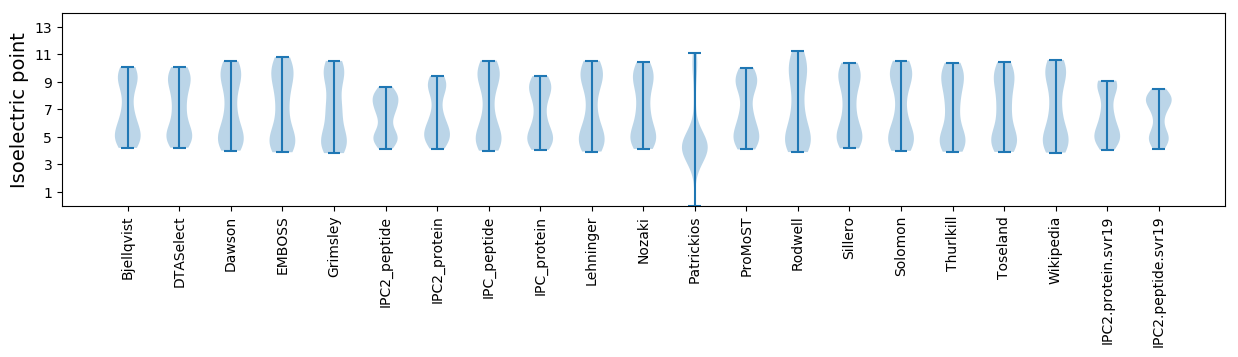
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
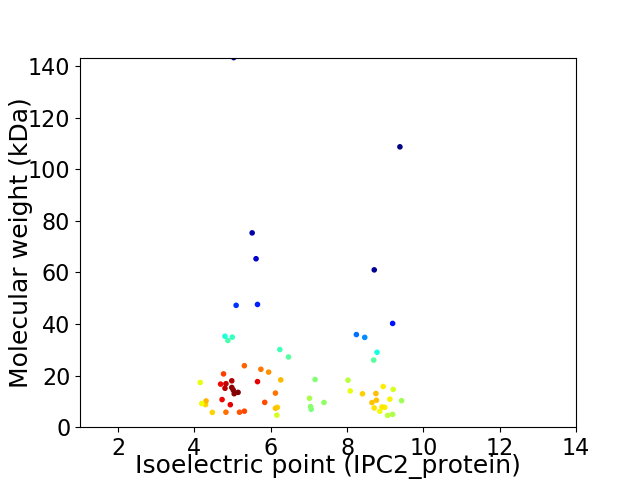
Virtual 2D-PAGE plot for 67 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345AQS5|A0A345AQS5_9CAUD Uncharacterized protein OS=Staphylococcus phage SA537ruMSSAST97 OX=2268623 PE=4 SV=1
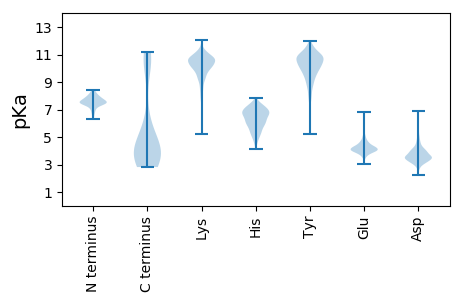
MM1 pKa = 7.42KK2 pKa = 10.25NYY4 pKa = 10.4YY5 pKa = 9.86HH6 pKa = 7.24LLSFDD11 pKa = 5.04DD12 pKa = 5.61DD13 pKa = 4.5LANDD17 pKa = 4.17AANDD21 pKa = 4.01LLKK24 pKa = 10.8EE25 pKa = 4.08GWDD28 pKa = 3.34IVHH31 pKa = 6.66VGTKK35 pKa = 9.51LVKK38 pKa = 10.36ILDD41 pKa = 3.92NGQAYY46 pKa = 10.82YY47 pKa = 8.91NTEE50 pKa = 3.97YY51 pKa = 11.47VLGGTKK57 pKa = 9.45NQYY60 pKa = 9.36EE61 pKa = 4.83KK62 pKa = 11.2YY63 pKa = 10.75LEE65 pKa = 4.8DD66 pKa = 4.04CQQSEE71 pKa = 4.31LDD73 pKa = 3.76YY74 pKa = 11.3FF75 pKa = 4.23
MM1 pKa = 7.42KK2 pKa = 10.25NYY4 pKa = 10.4YY5 pKa = 9.86HH6 pKa = 7.24LLSFDD11 pKa = 5.04DD12 pKa = 5.61DD13 pKa = 4.5LANDD17 pKa = 4.17AANDD21 pKa = 4.01LLKK24 pKa = 10.8EE25 pKa = 4.08GWDD28 pKa = 3.34IVHH31 pKa = 6.66VGTKK35 pKa = 9.51LVKK38 pKa = 10.36ILDD41 pKa = 3.92NGQAYY46 pKa = 10.82YY47 pKa = 8.91NTEE50 pKa = 3.97YY51 pKa = 11.47VLGGTKK57 pKa = 9.45NQYY60 pKa = 9.36EE61 pKa = 4.83KK62 pKa = 11.2YY63 pKa = 10.75LEE65 pKa = 4.8DD66 pKa = 4.04CQQSEE71 pKa = 4.31LDD73 pKa = 3.76YY74 pKa = 11.3FF75 pKa = 4.23
Molecular weight: 8.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345AQW8|A0A345AQW8_9CAUD Uncharacterized protein OS=Staphylococcus phage SA537ruMSSAST97 OX=2268623 PE=4 SV=1
MM1 pKa = 8.25PITLIVGALVGIGIAFYY18 pKa = 10.02QAYY21 pKa = 9.25KK22 pKa = 10.26RR23 pKa = 11.84SEE25 pKa = 3.95TFRR28 pKa = 11.84NIVNQAISGVANAFKK43 pKa = 10.38AAKK46 pKa = 9.72LALQGFFDD54 pKa = 5.0LFKK57 pKa = 10.77GDD59 pKa = 3.29SKK61 pKa = 11.62GAVTLEE67 pKa = 4.39KK68 pKa = 10.08IFPPEE73 pKa = 3.69TVAGIQNVVNTIRR86 pKa = 11.84TTFFKK91 pKa = 11.04VVDD94 pKa = 5.01AIVGFAKK101 pKa = 10.66EE102 pKa = 4.13IGAQLASFWKK112 pKa = 10.26EE113 pKa = 3.52NGSEE117 pKa = 3.87ITQALQNIAGFIKK130 pKa = 10.05ATFEE134 pKa = 4.49FIFNFIIKK142 pKa = 9.89PIMFAIWQVMQFIWPAVKK160 pKa = 10.39ALIVSTWEE168 pKa = 3.89NIKK171 pKa = 10.86GVIQGAINIILGIIKK186 pKa = 9.74VFSSLFTGNWRR197 pKa = 11.84GVWDD201 pKa = 5.1GIVMILKK208 pKa = 8.67GTVQLIWNLIQLWFIGKK225 pKa = 8.65ILGVVRR231 pKa = 11.84YY232 pKa = 9.14FGGLLKK238 pKa = 10.93GLITIIWVAIIGVFKK253 pKa = 10.88KK254 pKa = 10.63SLSAIWNATKK264 pKa = 10.51SIFGFLFNSVKK275 pKa = 10.81SIFTNMKK282 pKa = 9.13NWLSSTWNNIKK293 pKa = 10.89SNTVGKK299 pKa = 10.05AHH301 pKa = 6.79SLFTGVRR308 pKa = 11.84SKK310 pKa = 9.99FTSLWNATKK319 pKa = 10.52DD320 pKa = 2.9IFTKK324 pKa = 10.34LRR326 pKa = 11.84NWMSNIWNSIKK337 pKa = 10.99DD338 pKa = 3.66NTVGIAGRR346 pKa = 11.84LWDD349 pKa = 3.65RR350 pKa = 11.84VRR352 pKa = 11.84NIFGSMRR359 pKa = 11.84DD360 pKa = 3.59GLKK363 pKa = 10.75SIIGKK368 pKa = 9.69IKK370 pKa = 10.09DD371 pKa = 3.91HH372 pKa = 6.74IGGMVDD378 pKa = 3.0AVKK381 pKa = 10.61RR382 pKa = 11.84GLNKK386 pKa = 10.47LIEE389 pKa = 4.41GLNWVGGKK397 pKa = 10.27LGMDD401 pKa = 5.36KK402 pKa = 10.4IPKK405 pKa = 8.99LHH407 pKa = 6.73TGTEE411 pKa = 4.34HH412 pKa = 5.61THH414 pKa = 4.28TTTRR418 pKa = 11.84LVKK421 pKa = 10.0NGKK424 pKa = 8.59IARR427 pKa = 11.84DD428 pKa = 3.55TFATVGDD435 pKa = 3.94KK436 pKa = 11.29GRR438 pKa = 11.84GNGPNGFRR446 pKa = 11.84NEE448 pKa = 4.12MIEE451 pKa = 4.07FPNGKK456 pKa = 9.2RR457 pKa = 11.84VLTPNTDD464 pKa = 2.88TTAYY468 pKa = 9.76LPKK471 pKa = 10.41GSKK474 pKa = 9.93VYY476 pKa = 10.88NGAQTYY482 pKa = 11.36SMLNGTLPRR491 pKa = 11.84FSIGTMWKK499 pKa = 9.9DD500 pKa = 3.23IKK502 pKa = 10.95SGASSAFNWTKK513 pKa = 11.04DD514 pKa = 3.55QIGKK518 pKa = 6.62GTKK521 pKa = 8.59WLGDD525 pKa = 3.52KK526 pKa = 11.04VGDD529 pKa = 3.83VMDD532 pKa = 6.29FIDD535 pKa = 5.68NPGKK539 pKa = 10.35LLNYY543 pKa = 9.05VLQAFGVDD551 pKa = 4.58FSSLTKK557 pKa = 10.72GMGIAGDD564 pKa = 3.52ITKK567 pKa = 10.45AAWSKK572 pKa = 10.35IKK574 pKa = 10.52KK575 pKa = 9.78SAIKK579 pKa = 9.68WLEE582 pKa = 3.74DD583 pKa = 3.04AFAEE587 pKa = 4.44SGDD590 pKa = 3.93GGVLDD595 pKa = 4.17MNKK598 pKa = 9.58LRR600 pKa = 11.84YY601 pKa = 9.58LYY603 pKa = 10.75GHH605 pKa = 6.3TAAYY609 pKa = 8.63TRR611 pKa = 11.84EE612 pKa = 4.0TGRR615 pKa = 11.84PFHH618 pKa = 6.98EE619 pKa = 5.15GLDD622 pKa = 3.37FDD624 pKa = 5.43YY625 pKa = 10.49IYY627 pKa = 10.86EE628 pKa = 4.43PVPSTINGRR637 pKa = 11.84AQVMPFHH644 pKa = 6.69NGGYY648 pKa = 9.1GKK650 pKa = 9.26WVKK653 pKa = 9.66IVKK656 pKa = 9.85GALEE660 pKa = 4.22VIYY663 pKa = 10.74AHH665 pKa = 6.91LSKK668 pKa = 11.25YY669 pKa = 9.43KK670 pKa = 10.69VKK672 pKa = 9.65TGQQVRR678 pKa = 11.84VGQTVGISGNTGFSTGPHH696 pKa = 5.68LHH698 pKa = 7.05YY699 pKa = 10.8EE700 pKa = 4.1MRR702 pKa = 11.84WNGRR706 pKa = 11.84HH707 pKa = 6.33RR708 pKa = 11.84DD709 pKa = 3.7PLPWLRR715 pKa = 11.84KK716 pKa = 9.28NNGGGKK722 pKa = 8.03STPGGNGAANARR734 pKa = 11.84RR735 pKa = 11.84AIKK738 pKa = 10.02AAQNILGGRR747 pKa = 11.84YY748 pKa = 8.1KK749 pKa = 10.85ASWITNEE756 pKa = 3.8MMRR759 pKa = 11.84VASRR763 pKa = 11.84EE764 pKa = 4.02SNYY767 pKa = 9.28TANAVNNWDD776 pKa = 3.53SNARR780 pKa = 11.84AGIPSRR786 pKa = 11.84GMFQMIDD793 pKa = 3.41PSFRR797 pKa = 11.84AYY799 pKa = 10.42AKK801 pKa = 10.18SGYY804 pKa = 9.89NNPLNPTHH812 pKa = 6.01QAISAMRR819 pKa = 11.84YY820 pKa = 9.01IVGKK824 pKa = 8.54WVPRR828 pKa = 11.84TGSWRR833 pKa = 11.84AAFKK837 pKa = 10.7RR838 pKa = 11.84AGDD841 pKa = 3.57YY842 pKa = 10.87AYY844 pKa = 9.48ATGGKK849 pKa = 9.28VYY851 pKa = 10.84NGLYY855 pKa = 10.37HH856 pKa = 7.48LGEE859 pKa = 4.18EE860 pKa = 5.76GYY862 pKa = 9.66PEE864 pKa = 4.64WIIPTDD870 pKa = 3.52PSRR873 pKa = 11.84ANEE876 pKa = 3.92AHH878 pKa = 7.18KK879 pKa = 10.95LLALAANDD887 pKa = 3.04IDD889 pKa = 4.02NRR891 pKa = 11.84SKK893 pKa = 11.26NKK895 pKa = 10.15RR896 pKa = 11.84PNNLPNPSISNSDD909 pKa = 2.99TNYY912 pKa = 10.16IHH914 pKa = 6.64TLEE917 pKa = 4.51NKK919 pKa = 9.81LDD921 pKa = 3.73AVINCLVSLVEE932 pKa = 4.34SNQVIADD939 pKa = 3.63KK940 pKa = 10.76DD941 pKa = 3.8YY942 pKa = 10.92EE943 pKa = 4.23PVINKK948 pKa = 9.14YY949 pKa = 9.6VFEE952 pKa = 5.09DD953 pKa = 3.95EE954 pKa = 4.6VNNSIDD960 pKa = 3.0KK961 pKa = 10.58RR962 pKa = 11.84EE963 pKa = 3.96RR964 pKa = 11.84HH965 pKa = 5.3EE966 pKa = 4.02STRR969 pKa = 11.84VRR971 pKa = 11.84FRR973 pKa = 11.84RR974 pKa = 11.84GGTIII979 pKa = 4.08
MM1 pKa = 8.25PITLIVGALVGIGIAFYY18 pKa = 10.02QAYY21 pKa = 9.25KK22 pKa = 10.26RR23 pKa = 11.84SEE25 pKa = 3.95TFRR28 pKa = 11.84NIVNQAISGVANAFKK43 pKa = 10.38AAKK46 pKa = 9.72LALQGFFDD54 pKa = 5.0LFKK57 pKa = 10.77GDD59 pKa = 3.29SKK61 pKa = 11.62GAVTLEE67 pKa = 4.39KK68 pKa = 10.08IFPPEE73 pKa = 3.69TVAGIQNVVNTIRR86 pKa = 11.84TTFFKK91 pKa = 11.04VVDD94 pKa = 5.01AIVGFAKK101 pKa = 10.66EE102 pKa = 4.13IGAQLASFWKK112 pKa = 10.26EE113 pKa = 3.52NGSEE117 pKa = 3.87ITQALQNIAGFIKK130 pKa = 10.05ATFEE134 pKa = 4.49FIFNFIIKK142 pKa = 9.89PIMFAIWQVMQFIWPAVKK160 pKa = 10.39ALIVSTWEE168 pKa = 3.89NIKK171 pKa = 10.86GVIQGAINIILGIIKK186 pKa = 9.74VFSSLFTGNWRR197 pKa = 11.84GVWDD201 pKa = 5.1GIVMILKK208 pKa = 8.67GTVQLIWNLIQLWFIGKK225 pKa = 8.65ILGVVRR231 pKa = 11.84YY232 pKa = 9.14FGGLLKK238 pKa = 10.93GLITIIWVAIIGVFKK253 pKa = 10.88KK254 pKa = 10.63SLSAIWNATKK264 pKa = 10.51SIFGFLFNSVKK275 pKa = 10.81SIFTNMKK282 pKa = 9.13NWLSSTWNNIKK293 pKa = 10.89SNTVGKK299 pKa = 10.05AHH301 pKa = 6.79SLFTGVRR308 pKa = 11.84SKK310 pKa = 9.99FTSLWNATKK319 pKa = 10.52DD320 pKa = 2.9IFTKK324 pKa = 10.34LRR326 pKa = 11.84NWMSNIWNSIKK337 pKa = 10.99DD338 pKa = 3.66NTVGIAGRR346 pKa = 11.84LWDD349 pKa = 3.65RR350 pKa = 11.84VRR352 pKa = 11.84NIFGSMRR359 pKa = 11.84DD360 pKa = 3.59GLKK363 pKa = 10.75SIIGKK368 pKa = 9.69IKK370 pKa = 10.09DD371 pKa = 3.91HH372 pKa = 6.74IGGMVDD378 pKa = 3.0AVKK381 pKa = 10.61RR382 pKa = 11.84GLNKK386 pKa = 10.47LIEE389 pKa = 4.41GLNWVGGKK397 pKa = 10.27LGMDD401 pKa = 5.36KK402 pKa = 10.4IPKK405 pKa = 8.99LHH407 pKa = 6.73TGTEE411 pKa = 4.34HH412 pKa = 5.61THH414 pKa = 4.28TTTRR418 pKa = 11.84LVKK421 pKa = 10.0NGKK424 pKa = 8.59IARR427 pKa = 11.84DD428 pKa = 3.55TFATVGDD435 pKa = 3.94KK436 pKa = 11.29GRR438 pKa = 11.84GNGPNGFRR446 pKa = 11.84NEE448 pKa = 4.12MIEE451 pKa = 4.07FPNGKK456 pKa = 9.2RR457 pKa = 11.84VLTPNTDD464 pKa = 2.88TTAYY468 pKa = 9.76LPKK471 pKa = 10.41GSKK474 pKa = 9.93VYY476 pKa = 10.88NGAQTYY482 pKa = 11.36SMLNGTLPRR491 pKa = 11.84FSIGTMWKK499 pKa = 9.9DD500 pKa = 3.23IKK502 pKa = 10.95SGASSAFNWTKK513 pKa = 11.04DD514 pKa = 3.55QIGKK518 pKa = 6.62GTKK521 pKa = 8.59WLGDD525 pKa = 3.52KK526 pKa = 11.04VGDD529 pKa = 3.83VMDD532 pKa = 6.29FIDD535 pKa = 5.68NPGKK539 pKa = 10.35LLNYY543 pKa = 9.05VLQAFGVDD551 pKa = 4.58FSSLTKK557 pKa = 10.72GMGIAGDD564 pKa = 3.52ITKK567 pKa = 10.45AAWSKK572 pKa = 10.35IKK574 pKa = 10.52KK575 pKa = 9.78SAIKK579 pKa = 9.68WLEE582 pKa = 3.74DD583 pKa = 3.04AFAEE587 pKa = 4.44SGDD590 pKa = 3.93GGVLDD595 pKa = 4.17MNKK598 pKa = 9.58LRR600 pKa = 11.84YY601 pKa = 9.58LYY603 pKa = 10.75GHH605 pKa = 6.3TAAYY609 pKa = 8.63TRR611 pKa = 11.84EE612 pKa = 4.0TGRR615 pKa = 11.84PFHH618 pKa = 6.98EE619 pKa = 5.15GLDD622 pKa = 3.37FDD624 pKa = 5.43YY625 pKa = 10.49IYY627 pKa = 10.86EE628 pKa = 4.43PVPSTINGRR637 pKa = 11.84AQVMPFHH644 pKa = 6.69NGGYY648 pKa = 9.1GKK650 pKa = 9.26WVKK653 pKa = 9.66IVKK656 pKa = 9.85GALEE660 pKa = 4.22VIYY663 pKa = 10.74AHH665 pKa = 6.91LSKK668 pKa = 11.25YY669 pKa = 9.43KK670 pKa = 10.69VKK672 pKa = 9.65TGQQVRR678 pKa = 11.84VGQTVGISGNTGFSTGPHH696 pKa = 5.68LHH698 pKa = 7.05YY699 pKa = 10.8EE700 pKa = 4.1MRR702 pKa = 11.84WNGRR706 pKa = 11.84HH707 pKa = 6.33RR708 pKa = 11.84DD709 pKa = 3.7PLPWLRR715 pKa = 11.84KK716 pKa = 9.28NNGGGKK722 pKa = 8.03STPGGNGAANARR734 pKa = 11.84RR735 pKa = 11.84AIKK738 pKa = 10.02AAQNILGGRR747 pKa = 11.84YY748 pKa = 8.1KK749 pKa = 10.85ASWITNEE756 pKa = 3.8MMRR759 pKa = 11.84VASRR763 pKa = 11.84EE764 pKa = 4.02SNYY767 pKa = 9.28TANAVNNWDD776 pKa = 3.53SNARR780 pKa = 11.84AGIPSRR786 pKa = 11.84GMFQMIDD793 pKa = 3.41PSFRR797 pKa = 11.84AYY799 pKa = 10.42AKK801 pKa = 10.18SGYY804 pKa = 9.89NNPLNPTHH812 pKa = 6.01QAISAMRR819 pKa = 11.84YY820 pKa = 9.01IVGKK824 pKa = 8.54WVPRR828 pKa = 11.84TGSWRR833 pKa = 11.84AAFKK837 pKa = 10.7RR838 pKa = 11.84AGDD841 pKa = 3.57YY842 pKa = 10.87AYY844 pKa = 9.48ATGGKK849 pKa = 9.28VYY851 pKa = 10.84NGLYY855 pKa = 10.37HH856 pKa = 7.48LGEE859 pKa = 4.18EE860 pKa = 5.76GYY862 pKa = 9.66PEE864 pKa = 4.64WIIPTDD870 pKa = 3.52PSRR873 pKa = 11.84ANEE876 pKa = 3.92AHH878 pKa = 7.18KK879 pKa = 10.95LLALAANDD887 pKa = 3.04IDD889 pKa = 4.02NRR891 pKa = 11.84SKK893 pKa = 11.26NKK895 pKa = 10.15RR896 pKa = 11.84PNNLPNPSISNSDD909 pKa = 2.99TNYY912 pKa = 10.16IHH914 pKa = 6.64TLEE917 pKa = 4.51NKK919 pKa = 9.81LDD921 pKa = 3.73AVINCLVSLVEE932 pKa = 4.34SNQVIADD939 pKa = 3.63KK940 pKa = 10.76DD941 pKa = 3.8YY942 pKa = 10.92EE943 pKa = 4.23PVINKK948 pKa = 9.14YY949 pKa = 9.6VFEE952 pKa = 5.09DD953 pKa = 3.95EE954 pKa = 4.6VNNSIDD960 pKa = 3.0KK961 pKa = 10.58RR962 pKa = 11.84EE963 pKa = 3.96RR964 pKa = 11.84HH965 pKa = 5.3EE966 pKa = 4.02STRR969 pKa = 11.84VRR971 pKa = 11.84FRR973 pKa = 11.84RR974 pKa = 11.84GGTIII979 pKa = 4.08
Molecular weight: 108.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12850 |
40 |
1260 |
191.8 |
22.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.432 ± 0.334 | 0.56 ± 0.094 |
6.272 ± 0.32 | 7.922 ± 0.53 |
4.086 ± 0.236 | 5.362 ± 0.448 |
1.681 ± 0.151 | 8.016 ± 0.322 |
9.3 ± 0.327 | 8.14 ± 0.272 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.514 ± 0.238 | 6.911 ± 0.298 |
2.42 ± 0.176 | 3.977 ± 0.246 |
4.389 ± 0.198 | 5.977 ± 0.258 |
5.821 ± 0.215 | 5.844 ± 0.211 |
1.074 ± 0.19 | 4.304 ± 0.248 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
