
Pseudoclavibacter sp. RFBA6
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Pseudoclavibacter; unclassified Pseudoclavibacter
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
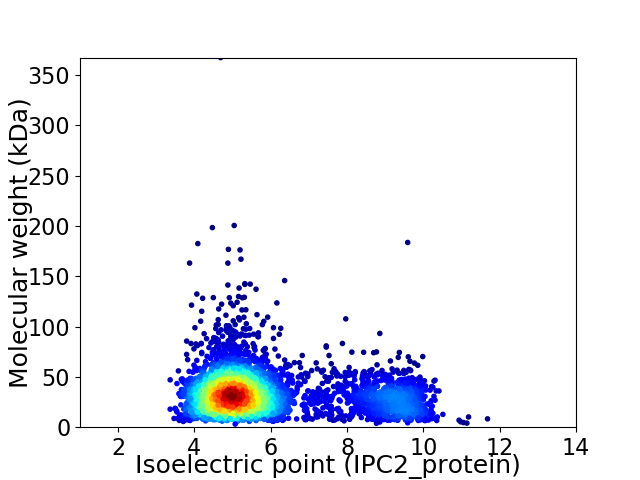
Virtual 2D-PAGE plot for 3544 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
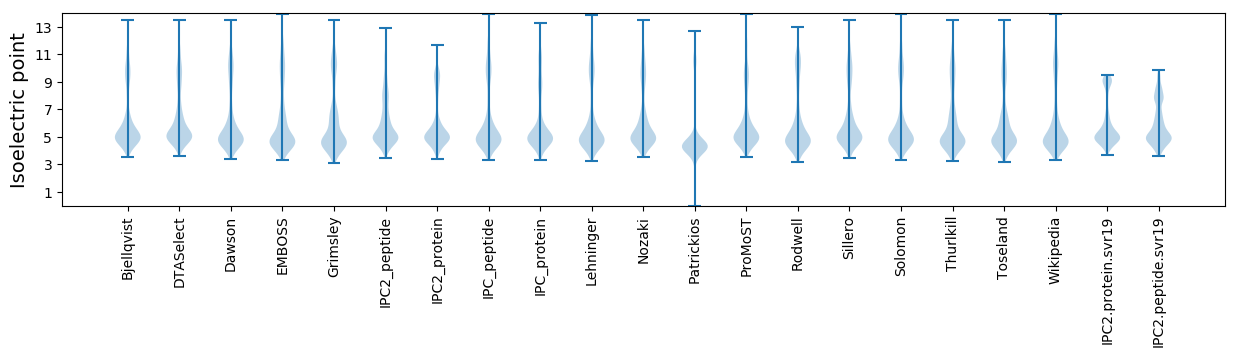
* You can choose from 21 different methods for calculating isoelectric point
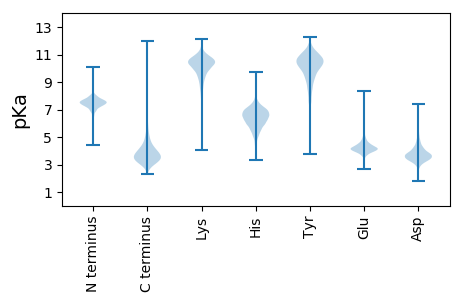
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S5X8Z1|A0A2S5X8Z1_9MICO Choline dehydrogenase OS=Pseudoclavibacter sp. RFBA6 OX=2080573 GN=C5C17_03740 PE=4 SV=1
MM1 pKa = 7.06SHH3 pKa = 6.61SRR5 pKa = 11.84NQRR8 pKa = 11.84MRR10 pKa = 11.84LSAALALPVAGVLALTGCASGGGDD34 pKa = 3.44TAGGGATEE42 pKa = 4.97FSLTYY47 pKa = 9.26ATSNNLEE54 pKa = 4.09NPYY57 pKa = 10.24EE58 pKa = 4.33ALATAYY64 pKa = 9.78MEE66 pKa = 5.05ANPDD70 pKa = 3.37VKK72 pKa = 9.98ITLNPQPNDD81 pKa = 3.21RR82 pKa = 11.84YY83 pKa = 11.08GEE85 pKa = 4.11TLRR88 pKa = 11.84TQLQAGNAPDD98 pKa = 4.48VIQTAPGSGQGQAVLSLGEE117 pKa = 4.29AGFLAPLGEE126 pKa = 4.35TASGVIPDD134 pKa = 4.18GSEE137 pKa = 3.52ALYY140 pKa = 9.67TYY142 pKa = 10.13EE143 pKa = 4.66DD144 pKa = 3.39QVLAQPVDD152 pKa = 3.87FTVAGFVYY160 pKa = 10.71SNASAEE166 pKa = 4.28AAGVTSYY173 pKa = 10.48PADD176 pKa = 3.54TAEE179 pKa = 5.08LLGKK183 pKa = 10.22CADD186 pKa = 3.84LTSQGKK192 pKa = 9.9SMIVIAGAAGPNTGMTAMSISATRR216 pKa = 11.84VYY218 pKa = 11.23AEE220 pKa = 3.98TPDD223 pKa = 3.5WNEE226 pKa = 3.04QRR228 pKa = 11.84AAGDD232 pKa = 3.32VTFADD237 pKa = 3.68SQGWKK242 pKa = 8.79DD243 pKa = 3.54TLEE246 pKa = 4.4TIIEE250 pKa = 4.31LKK252 pKa = 10.69DD253 pKa = 3.38AGCFQPGAEE262 pKa = 4.04GAGFDD267 pKa = 4.88VITNNITQGTSLGGFLPGSSANEE290 pKa = 3.66LAEE293 pKa = 4.65ANPDD297 pKa = 3.23LDD299 pKa = 4.6FSIEE303 pKa = 4.2PFPSADD309 pKa = 2.97GGKK312 pKa = 9.39PYY314 pKa = 11.2VLASSNYY321 pKa = 9.01SLSINAASEE330 pKa = 4.15KK331 pKa = 10.62QEE333 pKa = 3.88AAQAFLDD340 pKa = 3.95WVAGDD345 pKa = 3.87EE346 pKa = 4.31GAKK349 pKa = 10.42LFSDD353 pKa = 4.06VSGALPVSGIEE364 pKa = 4.0SYY366 pKa = 10.9EE367 pKa = 4.2FEE369 pKa = 4.47GSPYY373 pKa = 11.0ASVEE377 pKa = 4.01QVLTDD382 pKa = 3.59GSYY385 pKa = 10.03TPLPNSLWPNQAVYY399 pKa = 10.99DD400 pKa = 4.09EE401 pKa = 4.55LQKK404 pKa = 11.02GVQGLFTGQTTVDD417 pKa = 3.77SVLSAMDD424 pKa = 4.14AAWDD428 pKa = 3.69SS429 pKa = 3.6
MM1 pKa = 7.06SHH3 pKa = 6.61SRR5 pKa = 11.84NQRR8 pKa = 11.84MRR10 pKa = 11.84LSAALALPVAGVLALTGCASGGGDD34 pKa = 3.44TAGGGATEE42 pKa = 4.97FSLTYY47 pKa = 9.26ATSNNLEE54 pKa = 4.09NPYY57 pKa = 10.24EE58 pKa = 4.33ALATAYY64 pKa = 9.78MEE66 pKa = 5.05ANPDD70 pKa = 3.37VKK72 pKa = 9.98ITLNPQPNDD81 pKa = 3.21RR82 pKa = 11.84YY83 pKa = 11.08GEE85 pKa = 4.11TLRR88 pKa = 11.84TQLQAGNAPDD98 pKa = 4.48VIQTAPGSGQGQAVLSLGEE117 pKa = 4.29AGFLAPLGEE126 pKa = 4.35TASGVIPDD134 pKa = 4.18GSEE137 pKa = 3.52ALYY140 pKa = 9.67TYY142 pKa = 10.13EE143 pKa = 4.66DD144 pKa = 3.39QVLAQPVDD152 pKa = 3.87FTVAGFVYY160 pKa = 10.71SNASAEE166 pKa = 4.28AAGVTSYY173 pKa = 10.48PADD176 pKa = 3.54TAEE179 pKa = 5.08LLGKK183 pKa = 10.22CADD186 pKa = 3.84LTSQGKK192 pKa = 9.9SMIVIAGAAGPNTGMTAMSISATRR216 pKa = 11.84VYY218 pKa = 11.23AEE220 pKa = 3.98TPDD223 pKa = 3.5WNEE226 pKa = 3.04QRR228 pKa = 11.84AAGDD232 pKa = 3.32VTFADD237 pKa = 3.68SQGWKK242 pKa = 8.79DD243 pKa = 3.54TLEE246 pKa = 4.4TIIEE250 pKa = 4.31LKK252 pKa = 10.69DD253 pKa = 3.38AGCFQPGAEE262 pKa = 4.04GAGFDD267 pKa = 4.88VITNNITQGTSLGGFLPGSSANEE290 pKa = 3.66LAEE293 pKa = 4.65ANPDD297 pKa = 3.23LDD299 pKa = 4.6FSIEE303 pKa = 4.2PFPSADD309 pKa = 2.97GGKK312 pKa = 9.39PYY314 pKa = 11.2VLASSNYY321 pKa = 9.01SLSINAASEE330 pKa = 4.15KK331 pKa = 10.62QEE333 pKa = 3.88AAQAFLDD340 pKa = 3.95WVAGDD345 pKa = 3.87EE346 pKa = 4.31GAKK349 pKa = 10.42LFSDD353 pKa = 4.06VSGALPVSGIEE364 pKa = 4.0SYY366 pKa = 10.9EE367 pKa = 4.2FEE369 pKa = 4.47GSPYY373 pKa = 11.0ASVEE377 pKa = 4.01QVLTDD382 pKa = 3.59GSYY385 pKa = 10.03TPLPNSLWPNQAVYY399 pKa = 10.99DD400 pKa = 4.09EE401 pKa = 4.55LQKK404 pKa = 11.02GVQGLFTGQTTVDD417 pKa = 3.77SVLSAMDD424 pKa = 4.14AAWDD428 pKa = 3.69SS429 pKa = 3.6
Molecular weight: 44.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S5X5V3|A0A2S5X5V3_9MICO Amino acid ABC transporter permease OS=Pseudoclavibacter sp. RFBA6 OX=2080573 GN=C5C17_08645 PE=3 SV=1
MM1 pKa = 7.51PRR3 pKa = 11.84PRR5 pKa = 11.84RR6 pKa = 11.84GRR8 pKa = 11.84LRR10 pKa = 11.84RR11 pKa = 11.84LRR13 pKa = 11.84VRR15 pKa = 11.84SRR17 pKa = 11.84SRR19 pKa = 11.84RR20 pKa = 11.84STRR23 pKa = 11.84RR24 pKa = 11.84AGLQRR29 pKa = 11.84PRR31 pKa = 11.84RR32 pKa = 11.84LRR34 pKa = 11.84RR35 pKa = 11.84SAPRR39 pKa = 11.84GCRR42 pKa = 11.84RR43 pKa = 11.84PTGARR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84GRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84CPRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84EE61 pKa = 3.55PRR63 pKa = 11.84IRR65 pKa = 11.84TRR67 pKa = 11.84WRR69 pKa = 11.84SSRR72 pKa = 11.84LRR74 pKa = 11.84LRR76 pKa = 11.84IRR78 pKa = 11.84LRR80 pKa = 3.54
MM1 pKa = 7.51PRR3 pKa = 11.84PRR5 pKa = 11.84RR6 pKa = 11.84GRR8 pKa = 11.84LRR10 pKa = 11.84RR11 pKa = 11.84LRR13 pKa = 11.84VRR15 pKa = 11.84SRR17 pKa = 11.84SRR19 pKa = 11.84RR20 pKa = 11.84STRR23 pKa = 11.84RR24 pKa = 11.84AGLQRR29 pKa = 11.84PRR31 pKa = 11.84RR32 pKa = 11.84LRR34 pKa = 11.84RR35 pKa = 11.84SAPRR39 pKa = 11.84GCRR42 pKa = 11.84RR43 pKa = 11.84PTGARR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84GRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84CPRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84EE61 pKa = 3.55PRR63 pKa = 11.84IRR65 pKa = 11.84TRR67 pKa = 11.84WRR69 pKa = 11.84SSRR72 pKa = 11.84LRR74 pKa = 11.84LRR76 pKa = 11.84IRR78 pKa = 11.84LRR80 pKa = 3.54
Molecular weight: 10.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1157659 |
29 |
3465 |
326.7 |
34.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.114 ± 0.051 | 0.493 ± 0.008 |
5.855 ± 0.034 | 6.068 ± 0.034 |
3.281 ± 0.024 | 8.927 ± 0.034 |
1.951 ± 0.017 | 4.622 ± 0.035 |
2.11 ± 0.024 | 10.242 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.817 ± 0.016 | 2.086 ± 0.021 |
5.26 ± 0.031 | 2.992 ± 0.023 |
6.978 ± 0.043 | 6.191 ± 0.029 |
6.094 ± 0.032 | 8.619 ± 0.033 |
1.409 ± 0.018 | 1.891 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
