
Neodiprion sertifer nucleopolyhedrovirus
Taxonomy: Viruses; Naldaviricetes; Lefavirales; Baculoviridae; Gammabaculovirus
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
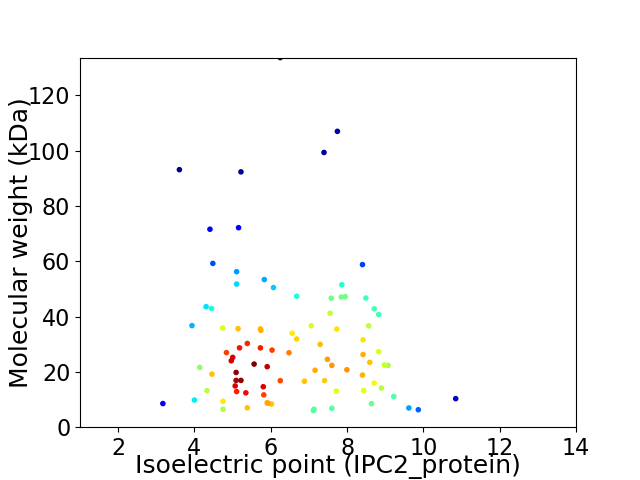
Virtual 2D-PAGE plot for 90 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
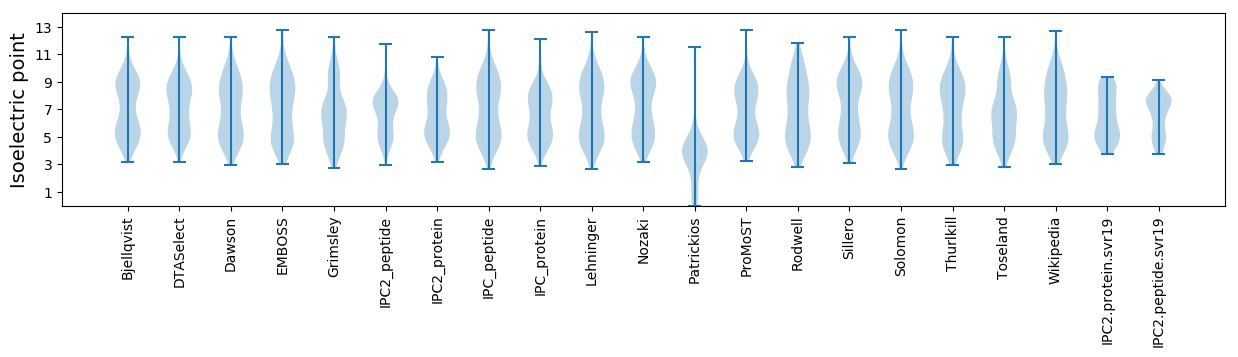
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6JKC3|Q6JKC3_9CBAC Uncharacterized protein OS=Neodiprion sertifer nucleopolyhedrovirus OX=111874 PE=4 SV=1
MM1 pKa = 7.65FSGLRR6 pKa = 11.84KK7 pKa = 7.27TAKK10 pKa = 10.12IYY12 pKa = 11.14DD13 pKa = 3.77NTADD17 pKa = 4.31LLVDD21 pKa = 3.47NTSLVVGKK29 pKa = 9.29FSNFDD34 pKa = 3.44AVFSLPSAKK43 pKa = 10.1SISKK47 pKa = 10.54GYY49 pKa = 10.02VIDD52 pKa = 3.65YY53 pKa = 7.98ATFIGNVDD61 pKa = 3.39TFSINKK67 pKa = 9.21ILRR70 pKa = 11.84QADD73 pKa = 3.63DD74 pKa = 3.79VNIEE78 pKa = 4.23TLFNATDD85 pKa = 3.46TDD87 pKa = 3.77IAGLNVLRR95 pKa = 11.84KK96 pKa = 9.31AANVPDD102 pKa = 3.46NTIYY106 pKa = 10.07VAEE109 pKa = 4.45VKK111 pKa = 10.4RR112 pKa = 11.84INLKK116 pKa = 10.27SLYY119 pKa = 9.63PSLDD123 pKa = 2.98VKK125 pKa = 10.37TYY127 pKa = 11.08DD128 pKa = 4.96GIASGLNNNPKK139 pKa = 10.2LYY141 pKa = 10.57SYY143 pKa = 11.42LKK145 pKa = 9.53GLGVATLAGGAVVLVLLGIDD165 pKa = 4.28LVQDD169 pKa = 4.39IIDD172 pKa = 3.82ALNRR176 pKa = 11.84TGGSYY181 pKa = 8.39FTYY184 pKa = 10.97GEE186 pKa = 4.05ADD188 pKa = 3.48NVEE191 pKa = 4.32SCYY194 pKa = 10.78LRR196 pKa = 11.84YY197 pKa = 9.74RR198 pKa = 11.84SCGVDD203 pKa = 3.2QSSVDD208 pKa = 3.39TTTYY212 pKa = 10.02CQNFLDD218 pKa = 5.83PILEE222 pKa = 4.61DD223 pKa = 4.57DD224 pKa = 4.1VTALTAICDD233 pKa = 4.07GYY235 pKa = 11.29DD236 pKa = 3.09IDD238 pKa = 5.06TEE240 pKa = 4.17ISVCRR245 pKa = 11.84QSDD248 pKa = 4.33PYY250 pKa = 11.27ADD252 pKa = 4.48PDD254 pKa = 3.74SEE256 pKa = 4.18QWVDD260 pKa = 3.5VSEE263 pKa = 4.18LAEE266 pKa = 4.21NQTLSCVEE274 pKa = 4.62PYY276 pKa = 10.0TFSDD280 pKa = 4.78LISDD284 pKa = 5.54LGLDD288 pKa = 3.51WLLGDD293 pKa = 5.76DD294 pKa = 4.91SILGNSFTSGSDD306 pKa = 3.6SISSLSSYY314 pKa = 9.83IGYY317 pKa = 9.39IAIFAIIIIVGIVFIKK333 pKa = 10.6LSKK336 pKa = 10.51AVSS339 pKa = 3.24
MM1 pKa = 7.65FSGLRR6 pKa = 11.84KK7 pKa = 7.27TAKK10 pKa = 10.12IYY12 pKa = 11.14DD13 pKa = 3.77NTADD17 pKa = 4.31LLVDD21 pKa = 3.47NTSLVVGKK29 pKa = 9.29FSNFDD34 pKa = 3.44AVFSLPSAKK43 pKa = 10.1SISKK47 pKa = 10.54GYY49 pKa = 10.02VIDD52 pKa = 3.65YY53 pKa = 7.98ATFIGNVDD61 pKa = 3.39TFSINKK67 pKa = 9.21ILRR70 pKa = 11.84QADD73 pKa = 3.63DD74 pKa = 3.79VNIEE78 pKa = 4.23TLFNATDD85 pKa = 3.46TDD87 pKa = 3.77IAGLNVLRR95 pKa = 11.84KK96 pKa = 9.31AANVPDD102 pKa = 3.46NTIYY106 pKa = 10.07VAEE109 pKa = 4.45VKK111 pKa = 10.4RR112 pKa = 11.84INLKK116 pKa = 10.27SLYY119 pKa = 9.63PSLDD123 pKa = 2.98VKK125 pKa = 10.37TYY127 pKa = 11.08DD128 pKa = 4.96GIASGLNNNPKK139 pKa = 10.2LYY141 pKa = 10.57SYY143 pKa = 11.42LKK145 pKa = 9.53GLGVATLAGGAVVLVLLGIDD165 pKa = 4.28LVQDD169 pKa = 4.39IIDD172 pKa = 3.82ALNRR176 pKa = 11.84TGGSYY181 pKa = 8.39FTYY184 pKa = 10.97GEE186 pKa = 4.05ADD188 pKa = 3.48NVEE191 pKa = 4.32SCYY194 pKa = 10.78LRR196 pKa = 11.84YY197 pKa = 9.74RR198 pKa = 11.84SCGVDD203 pKa = 3.2QSSVDD208 pKa = 3.39TTTYY212 pKa = 10.02CQNFLDD218 pKa = 5.83PILEE222 pKa = 4.61DD223 pKa = 4.57DD224 pKa = 4.1VTALTAICDD233 pKa = 4.07GYY235 pKa = 11.29DD236 pKa = 3.09IDD238 pKa = 5.06TEE240 pKa = 4.17ISVCRR245 pKa = 11.84QSDD248 pKa = 4.33PYY250 pKa = 11.27ADD252 pKa = 4.48PDD254 pKa = 3.74SEE256 pKa = 4.18QWVDD260 pKa = 3.5VSEE263 pKa = 4.18LAEE266 pKa = 4.21NQTLSCVEE274 pKa = 4.62PYY276 pKa = 10.0TFSDD280 pKa = 4.78LISDD284 pKa = 5.54LGLDD288 pKa = 3.51WLLGDD293 pKa = 5.76DD294 pKa = 4.91SILGNSFTSGSDD306 pKa = 3.6SISSLSSYY314 pKa = 9.83IGYY317 pKa = 9.39IAIFAIIIIVGIVFIKK333 pKa = 10.6LSKK336 pKa = 10.51AVSS339 pKa = 3.24
Molecular weight: 36.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6JKE7|Q6JKE7_9CBAC Uncharacterized protein OS=Neodiprion sertifer nucleopolyhedrovirus OX=111874 PE=4 SV=1
MM1 pKa = 7.62SDD3 pKa = 3.25SPVFNRR9 pKa = 11.84SSSRR13 pKa = 11.84NRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84FNLSLPRR24 pKa = 11.84AQVVLQRR31 pKa = 11.84IDD33 pKa = 3.82DD34 pKa = 4.1MPDD37 pKa = 2.58IMALDD42 pKa = 3.85HH43 pKa = 7.33QEE45 pKa = 3.84LNQRR49 pKa = 11.84KK50 pKa = 8.96SLIFYY55 pKa = 9.1LFKK58 pKa = 11.08
MM1 pKa = 7.62SDD3 pKa = 3.25SPVFNRR9 pKa = 11.84SSSRR13 pKa = 11.84NRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84FNLSLPRR24 pKa = 11.84AQVVLQRR31 pKa = 11.84IDD33 pKa = 3.82DD34 pKa = 4.1MPDD37 pKa = 2.58IMALDD42 pKa = 3.85HH43 pKa = 7.33QEE45 pKa = 3.84LNQRR49 pKa = 11.84KK50 pKa = 8.96SLIFYY55 pKa = 9.1LFKK58 pKa = 11.08
Molecular weight: 6.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
24263 |
54 |
1143 |
269.6 |
31.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.446 ± 0.196 | 2.774 ± 0.182 |
6.669 ± 0.245 | 5.185 ± 0.189 |
4.958 ± 0.193 | 2.864 ± 0.193 |
2.506 ± 0.173 | 9.245 ± 0.244 |
7.237 ± 0.326 | 8.68 ± 0.226 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.555 ± 0.135 | 8.305 ± 0.209 |
2.745 ± 0.135 | 3.681 ± 0.138 |
3.821 ± 0.196 | 7.089 ± 0.27 |
6.545 ± 0.28 | 6.075 ± 0.2 |
0.717 ± 0.06 | 4.905 ± 0.154 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
