
Bat hepatitis virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Blubervirales; Hepadnaviridae; unclassified Hepadnaviridae
Average proteome isoelectric point is 8.6
Get precalculated fractions of proteins
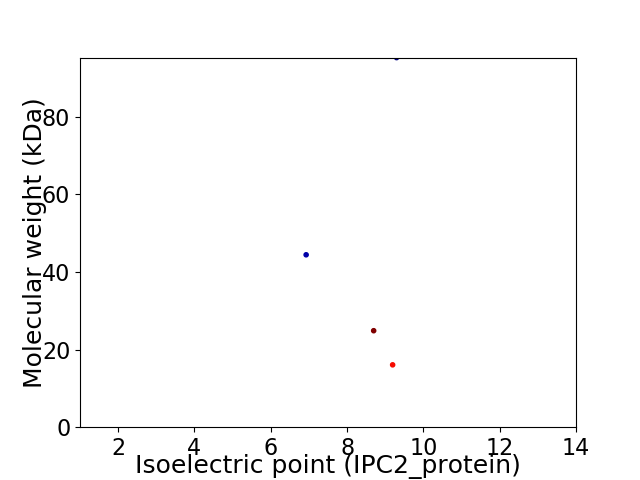
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
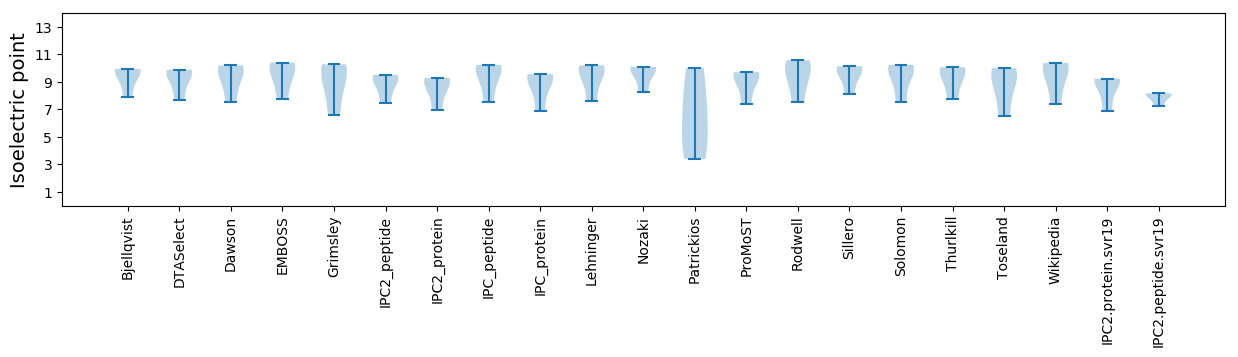
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M4PW60|M4PW60_9HEPA Core antigen OS=Bat hepatitis virus OX=1303386 PE=4 SV=1
MM1 pKa = 6.67GQNWSIPNPLGFLPEE16 pKa = 4.11HH17 pKa = 6.06QLPNPYY23 pKa = 10.64APVYY27 pKa = 10.26QDD29 pKa = 2.8WDD31 pKa = 3.94LNQDD35 pKa = 3.24KK36 pKa = 10.6DD37 pKa = 3.28QWPQARR43 pKa = 11.84EE44 pKa = 4.02VSPGAYY50 pKa = 9.78GLGFVPPHH58 pKa = 6.61GGLTGNLPFAQEE70 pKa = 4.35GNLTSNNPEE79 pKa = 4.08TIVYY83 pKa = 7.86QIPKK87 pKa = 10.37AKK89 pKa = 10.3DD90 pKa = 3.17EE91 pKa = 4.43SQTQVIQLPAPPRR104 pKa = 11.84TTTNRR109 pKa = 11.84KK110 pKa = 9.38KK111 pKa = 10.63GRR113 pKa = 11.84QPTPPTPPVRR123 pKa = 11.84VTHH126 pKa = 6.2PHH128 pKa = 6.5LNMPSWRR135 pKa = 11.84EE136 pKa = 3.88KK137 pKa = 10.16YY138 pKa = 10.45GHH140 pKa = 7.76LITAGGSSSKK150 pKa = 10.71LQIPAVTAASVTSSASSTIGDD171 pKa = 4.15PATDD175 pKa = 3.34MGITTSGFLVPLAGLQVGCFLLTKK199 pKa = 9.8ILEE202 pKa = 4.36IGKK205 pKa = 9.55SLDD208 pKa = 3.21WWWTSLSFPGGIPVCIGQNSQSQTSSHH235 pKa = 6.51SPTSCPPTCTGFRR248 pKa = 11.84WMWLRR253 pKa = 11.84RR254 pKa = 11.84FIIYY258 pKa = 10.48LLVLLLCAIFLLVLLDD274 pKa = 3.37WRR276 pKa = 11.84GFLPVCPLPTSSSTTTRR293 pKa = 11.84CSTCTATATDD303 pKa = 4.02QITWPYY309 pKa = 8.83CCCSKK314 pKa = 9.6PTDD317 pKa = 4.21GNCTCWPIPTSWALGKK333 pKa = 9.6FLWGWASARR342 pKa = 11.84FSWLNSLLPWLLWFAEE358 pKa = 4.29LSPTVWLLLIWMMWFWGPSLLTIFEE383 pKa = 4.43PFIPLCALFFVIWEE397 pKa = 4.24LWW399 pKa = 3.15
MM1 pKa = 6.67GQNWSIPNPLGFLPEE16 pKa = 4.11HH17 pKa = 6.06QLPNPYY23 pKa = 10.64APVYY27 pKa = 10.26QDD29 pKa = 2.8WDD31 pKa = 3.94LNQDD35 pKa = 3.24KK36 pKa = 10.6DD37 pKa = 3.28QWPQARR43 pKa = 11.84EE44 pKa = 4.02VSPGAYY50 pKa = 9.78GLGFVPPHH58 pKa = 6.61GGLTGNLPFAQEE70 pKa = 4.35GNLTSNNPEE79 pKa = 4.08TIVYY83 pKa = 7.86QIPKK87 pKa = 10.37AKK89 pKa = 10.3DD90 pKa = 3.17EE91 pKa = 4.43SQTQVIQLPAPPRR104 pKa = 11.84TTTNRR109 pKa = 11.84KK110 pKa = 9.38KK111 pKa = 10.63GRR113 pKa = 11.84QPTPPTPPVRR123 pKa = 11.84VTHH126 pKa = 6.2PHH128 pKa = 6.5LNMPSWRR135 pKa = 11.84EE136 pKa = 3.88KK137 pKa = 10.16YY138 pKa = 10.45GHH140 pKa = 7.76LITAGGSSSKK150 pKa = 10.71LQIPAVTAASVTSSASSTIGDD171 pKa = 4.15PATDD175 pKa = 3.34MGITTSGFLVPLAGLQVGCFLLTKK199 pKa = 9.8ILEE202 pKa = 4.36IGKK205 pKa = 9.55SLDD208 pKa = 3.21WWWTSLSFPGGIPVCIGQNSQSQTSSHH235 pKa = 6.51SPTSCPPTCTGFRR248 pKa = 11.84WMWLRR253 pKa = 11.84RR254 pKa = 11.84FIIYY258 pKa = 10.48LLVLLLCAIFLLVLLDD274 pKa = 3.37WRR276 pKa = 11.84GFLPVCPLPTSSSTTTRR293 pKa = 11.84CSTCTATATDD303 pKa = 4.02QITWPYY309 pKa = 8.83CCCSKK314 pKa = 9.6PTDD317 pKa = 4.21GNCTCWPIPTSWALGKK333 pKa = 9.6FLWGWASARR342 pKa = 11.84FSWLNSLLPWLLWFAEE358 pKa = 4.29LSPTVWLLLIWMMWFWGPSLLTIFEE383 pKa = 4.43PFIPLCALFFVIWEE397 pKa = 4.24LWW399 pKa = 3.15
Molecular weight: 44.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M4PV98|M4PV98_9HEPA Large envelope protein OS=Bat hepatitis virus OX=1303386 PE=4 SV=1
MM1 pKa = 7.69AARR4 pKa = 11.84LRR6 pKa = 11.84CQLDD10 pKa = 3.04ASGNVVLLRR19 pKa = 11.84PLCIEE24 pKa = 3.86SGGRR28 pKa = 11.84PVARR32 pKa = 11.84YY33 pKa = 8.84ARR35 pKa = 11.84HH36 pKa = 5.42SRR38 pKa = 11.84RR39 pKa = 11.84AAASPVPTVHH49 pKa = 6.48GPHH52 pKa = 4.82VTLRR56 pKa = 11.84RR57 pKa = 11.84LPICAASPAGPCALRR72 pKa = 11.84FTCANFSGCMEE83 pKa = 3.98TTMNFVTWLGTRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84TGAKK101 pKa = 9.76LNSGHH106 pKa = 6.22WDD108 pKa = 3.18MYY110 pKa = 10.64FRR112 pKa = 11.84EE113 pKa = 4.34SLMKK117 pKa = 10.13EE118 pKa = 3.78WEE120 pKa = 4.29EE121 pKa = 4.01KK122 pKa = 11.29GNDD125 pKa = 3.25QRR127 pKa = 11.84LYY129 pKa = 8.51TYY131 pKa = 10.93VSGGCRR137 pKa = 11.84HH138 pKa = 6.57KK139 pKa = 10.92LLCTLL144 pKa = 4.45
MM1 pKa = 7.69AARR4 pKa = 11.84LRR6 pKa = 11.84CQLDD10 pKa = 3.04ASGNVVLLRR19 pKa = 11.84PLCIEE24 pKa = 3.86SGGRR28 pKa = 11.84PVARR32 pKa = 11.84YY33 pKa = 8.84ARR35 pKa = 11.84HH36 pKa = 5.42SRR38 pKa = 11.84RR39 pKa = 11.84AAASPVPTVHH49 pKa = 6.48GPHH52 pKa = 4.82VTLRR56 pKa = 11.84RR57 pKa = 11.84LPICAASPAGPCALRR72 pKa = 11.84FTCANFSGCMEE83 pKa = 3.98TTMNFVTWLGTRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84TGAKK101 pKa = 9.76LNSGHH106 pKa = 6.22WDD108 pKa = 3.18MYY110 pKa = 10.64FRR112 pKa = 11.84EE113 pKa = 4.34SLMKK117 pKa = 10.13EE118 pKa = 3.78WEE120 pKa = 4.29EE121 pKa = 4.01KK122 pKa = 11.29GNDD125 pKa = 3.25QRR127 pKa = 11.84LYY129 pKa = 8.51TYY131 pKa = 10.93VSGGCRR137 pKa = 11.84HH138 pKa = 6.57KK139 pKa = 10.92LLCTLL144 pKa = 4.45
Molecular weight: 16.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1613 |
144 |
853 |
403.3 |
45.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.51 ± 0.58 | 3.038 ± 0.597 |
2.914 ± 0.21 | 2.976 ± 0.647 |
4.278 ± 0.229 | 6.696 ± 0.433 |
3.782 ± 0.809 | 3.968 ± 0.44 |
3.472 ± 0.853 | 11.841 ± 0.453 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.736 ± 0.269 | 3.472 ± 0.283 |
9.299 ± 0.803 | 3.658 ± 0.353 |
6.696 ± 1.568 | 8.493 ± 0.396 |
6.944 ± 0.959 | 4.65 ± 0.152 |
3.348 ± 0.728 | 2.232 ± 0.157 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
