
Duck faeces associated circular DNA virus 3
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.76
Get precalculated fractions of proteins
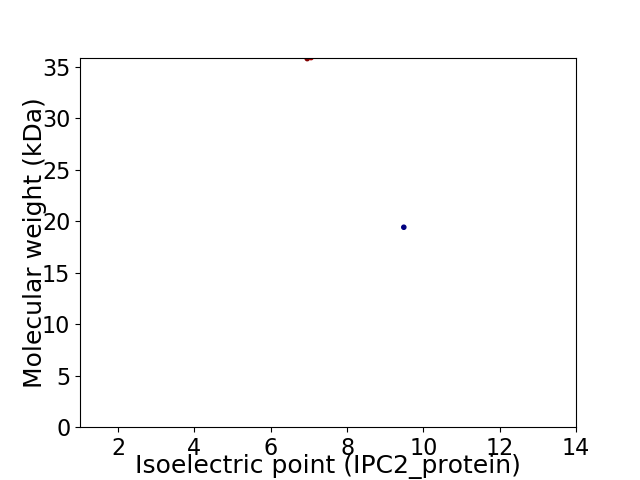
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A168MFT2|A0A168MFT2_9VIRU Replication associated protein OS=Duck faeces associated circular DNA virus 3 OX=1843770 PE=3 SV=1
MM1 pKa = 7.49EE2 pKa = 4.81HH3 pKa = 6.44HH4 pKa = 5.92QQIIQTPAIYY14 pKa = 10.17EE15 pKa = 4.07FRR17 pKa = 11.84GVRR20 pKa = 11.84ALITYY25 pKa = 7.42PQCNDD30 pKa = 3.02GNKK33 pKa = 10.2EE34 pKa = 3.89EE35 pKa = 5.35LLTFLKK41 pKa = 9.95TIYY44 pKa = 10.12NNKK47 pKa = 8.12IDD49 pKa = 4.21YY50 pKa = 10.55IVVCKK55 pKa = 10.03EE56 pKa = 3.82DD57 pKa = 3.15HH58 pKa = 6.72HH59 pKa = 6.18EE60 pKa = 4.11TDD62 pKa = 3.53GEE64 pKa = 4.47HH65 pKa = 4.54YY66 pKa = 10.09HH67 pKa = 7.77AYY69 pKa = 9.84IKK71 pKa = 10.68FSTRR75 pKa = 11.84AVVNANHH82 pKa = 5.88LTYY85 pKa = 10.6RR86 pKa = 11.84GIRR89 pKa = 11.84PNLEE93 pKa = 4.19KK94 pKa = 10.96VQSTAWKK101 pKa = 9.13AVRR104 pKa = 11.84YY105 pKa = 8.81VKK107 pKa = 10.49KK108 pKa = 10.31DD109 pKa = 3.21NNYY112 pKa = 9.51IEE114 pKa = 5.57EE115 pKa = 4.55GTCTDD120 pKa = 3.48VQKK123 pKa = 10.3LTTQEE128 pKa = 3.77KK129 pKa = 9.94YY130 pKa = 10.93KK131 pKa = 10.64LIKK134 pKa = 9.58EE135 pKa = 3.94KK136 pKa = 10.16TYY138 pKa = 10.73IEE140 pKa = 4.16IFEE143 pKa = 4.41MATLSLPEE151 pKa = 4.11LCKK154 pKa = 10.7VKK156 pKa = 10.46QIQKK160 pKa = 10.19EE161 pKa = 4.2LIVNDD166 pKa = 3.35WPMNGSKK173 pKa = 10.2NRR175 pKa = 11.84EE176 pKa = 4.14VYY178 pKa = 8.89WFYY181 pKa = 11.54GPTGTGKK188 pKa = 7.74TRR190 pKa = 11.84KK191 pKa = 7.78ATEE194 pKa = 3.95MMIQLYY200 pKa = 9.04GEE202 pKa = 4.24KK203 pKa = 9.48WVSLTGNLRR212 pKa = 11.84TFFDD216 pKa = 4.61PYY218 pKa = 10.64NGEE221 pKa = 3.97KK222 pKa = 10.7GVIFDD227 pKa = 5.72DD228 pKa = 3.36IRR230 pKa = 11.84KK231 pKa = 9.96GSIEE235 pKa = 4.36WNTLLTITDD244 pKa = 4.2RR245 pKa = 11.84YY246 pKa = 7.67RR247 pKa = 11.84TSVNVKK253 pKa = 9.63GSRR256 pKa = 11.84IPWLAEE262 pKa = 3.81TIIFTSPQHH271 pKa = 6.01FNDD274 pKa = 3.09VFTTEE279 pKa = 4.1KK280 pKa = 10.72DD281 pKa = 3.32GEE283 pKa = 4.12RR284 pKa = 11.84AQWDD288 pKa = 4.27GIEE291 pKa = 4.0QFEE294 pKa = 4.42RR295 pKa = 11.84RR296 pKa = 11.84IKK298 pKa = 9.81EE299 pKa = 3.91VKK301 pKa = 9.94EE302 pKa = 4.08FNN304 pKa = 3.56
MM1 pKa = 7.49EE2 pKa = 4.81HH3 pKa = 6.44HH4 pKa = 5.92QQIIQTPAIYY14 pKa = 10.17EE15 pKa = 4.07FRR17 pKa = 11.84GVRR20 pKa = 11.84ALITYY25 pKa = 7.42PQCNDD30 pKa = 3.02GNKK33 pKa = 10.2EE34 pKa = 3.89EE35 pKa = 5.35LLTFLKK41 pKa = 9.95TIYY44 pKa = 10.12NNKK47 pKa = 8.12IDD49 pKa = 4.21YY50 pKa = 10.55IVVCKK55 pKa = 10.03EE56 pKa = 3.82DD57 pKa = 3.15HH58 pKa = 6.72HH59 pKa = 6.18EE60 pKa = 4.11TDD62 pKa = 3.53GEE64 pKa = 4.47HH65 pKa = 4.54YY66 pKa = 10.09HH67 pKa = 7.77AYY69 pKa = 9.84IKK71 pKa = 10.68FSTRR75 pKa = 11.84AVVNANHH82 pKa = 5.88LTYY85 pKa = 10.6RR86 pKa = 11.84GIRR89 pKa = 11.84PNLEE93 pKa = 4.19KK94 pKa = 10.96VQSTAWKK101 pKa = 9.13AVRR104 pKa = 11.84YY105 pKa = 8.81VKK107 pKa = 10.49KK108 pKa = 10.31DD109 pKa = 3.21NNYY112 pKa = 9.51IEE114 pKa = 5.57EE115 pKa = 4.55GTCTDD120 pKa = 3.48VQKK123 pKa = 10.3LTTQEE128 pKa = 3.77KK129 pKa = 9.94YY130 pKa = 10.93KK131 pKa = 10.64LIKK134 pKa = 9.58EE135 pKa = 3.94KK136 pKa = 10.16TYY138 pKa = 10.73IEE140 pKa = 4.16IFEE143 pKa = 4.41MATLSLPEE151 pKa = 4.11LCKK154 pKa = 10.7VKK156 pKa = 10.46QIQKK160 pKa = 10.19EE161 pKa = 4.2LIVNDD166 pKa = 3.35WPMNGSKK173 pKa = 10.2NRR175 pKa = 11.84EE176 pKa = 4.14VYY178 pKa = 8.89WFYY181 pKa = 11.54GPTGTGKK188 pKa = 7.74TRR190 pKa = 11.84KK191 pKa = 7.78ATEE194 pKa = 3.95MMIQLYY200 pKa = 9.04GEE202 pKa = 4.24KK203 pKa = 9.48WVSLTGNLRR212 pKa = 11.84TFFDD216 pKa = 4.61PYY218 pKa = 10.64NGEE221 pKa = 3.97KK222 pKa = 10.7GVIFDD227 pKa = 5.72DD228 pKa = 3.36IRR230 pKa = 11.84KK231 pKa = 9.96GSIEE235 pKa = 4.36WNTLLTITDD244 pKa = 4.2RR245 pKa = 11.84YY246 pKa = 7.67RR247 pKa = 11.84TSVNVKK253 pKa = 9.63GSRR256 pKa = 11.84IPWLAEE262 pKa = 3.81TIIFTSPQHH271 pKa = 6.01FNDD274 pKa = 3.09VFTTEE279 pKa = 4.1KK280 pKa = 10.72DD281 pKa = 3.32GEE283 pKa = 4.12RR284 pKa = 11.84AQWDD288 pKa = 4.27GIEE291 pKa = 4.0QFEE294 pKa = 4.42RR295 pKa = 11.84RR296 pKa = 11.84IKK298 pKa = 9.81EE299 pKa = 3.91VKK301 pKa = 9.94EE302 pKa = 4.08FNN304 pKa = 3.56
Molecular weight: 35.81 kDa
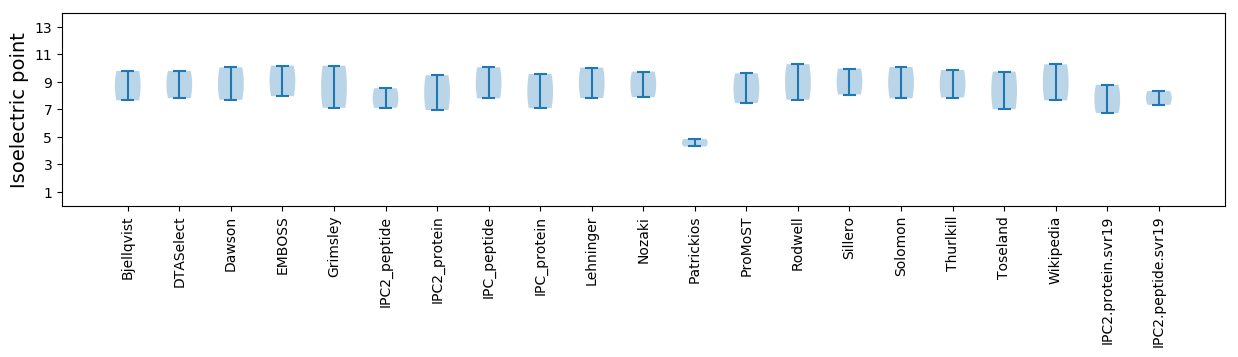
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168MFT2|A0A168MFT2_9VIRU Replication associated protein OS=Duck faeces associated circular DNA virus 3 OX=1843770 PE=3 SV=1
MM1 pKa = 7.25ARR3 pKa = 11.84YY4 pKa = 8.33YY5 pKa = 10.67KK6 pKa = 10.24YY7 pKa = 10.09RR8 pKa = 11.84RR9 pKa = 11.84VYY11 pKa = 10.33KK12 pKa = 10.16KK13 pKa = 10.18VYY15 pKa = 8.72PRR17 pKa = 11.84KK18 pKa = 8.66RR19 pKa = 11.84WASNIVTKK27 pKa = 10.85NVLLTVPANEE37 pKa = 3.82KK38 pKa = 9.4TVFSFSTLVANSAQTVTPTPTLLKK62 pKa = 10.48FGRR65 pKa = 11.84CKK67 pKa = 10.29IKK69 pKa = 10.73GDD71 pKa = 3.05IRR73 pKa = 11.84TDD75 pKa = 3.21VASEE79 pKa = 3.81NNYY82 pKa = 9.92VSGIMYY88 pKa = 9.34VIYY91 pKa = 10.46VPEE94 pKa = 4.53GFAVSPTLISQHH106 pKa = 6.24PEE108 pKa = 3.85YY109 pKa = 10.51IIGWTQISFDD119 pKa = 3.88SGNTFSFSSSLKK131 pKa = 10.23RR132 pKa = 11.84NLNSGDD138 pKa = 3.93RR139 pKa = 11.84IDD141 pKa = 5.5LFFSVDD147 pKa = 3.51SVNSVSAVRR156 pKa = 11.84NFNMYY161 pKa = 8.39FTAQYY166 pKa = 6.57WTSSAA171 pKa = 3.62
MM1 pKa = 7.25ARR3 pKa = 11.84YY4 pKa = 8.33YY5 pKa = 10.67KK6 pKa = 10.24YY7 pKa = 10.09RR8 pKa = 11.84RR9 pKa = 11.84VYY11 pKa = 10.33KK12 pKa = 10.16KK13 pKa = 10.18VYY15 pKa = 8.72PRR17 pKa = 11.84KK18 pKa = 8.66RR19 pKa = 11.84WASNIVTKK27 pKa = 10.85NVLLTVPANEE37 pKa = 3.82KK38 pKa = 9.4TVFSFSTLVANSAQTVTPTPTLLKK62 pKa = 10.48FGRR65 pKa = 11.84CKK67 pKa = 10.29IKK69 pKa = 10.73GDD71 pKa = 3.05IRR73 pKa = 11.84TDD75 pKa = 3.21VASEE79 pKa = 3.81NNYY82 pKa = 9.92VSGIMYY88 pKa = 9.34VIYY91 pKa = 10.46VPEE94 pKa = 4.53GFAVSPTLISQHH106 pKa = 6.24PEE108 pKa = 3.85YY109 pKa = 10.51IIGWTQISFDD119 pKa = 3.88SGNTFSFSSSLKK131 pKa = 10.23RR132 pKa = 11.84NLNSGDD138 pKa = 3.93RR139 pKa = 11.84IDD141 pKa = 5.5LFFSVDD147 pKa = 3.51SVNSVSAVRR156 pKa = 11.84NFNMYY161 pKa = 8.39FTAQYY166 pKa = 6.57WTSSAA171 pKa = 3.62
Molecular weight: 19.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
475 |
171 |
304 |
237.5 |
27.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.421 ± 0.799 | 1.053 ± 0.262 |
4.211 ± 0.393 | 6.737 ± 2.462 |
5.053 ± 0.773 | 5.053 ± 0.537 |
1.895 ± 0.733 | 7.368 ± 0.851 |
7.789 ± 1.087 | 5.684 ± 0.236 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.684 ± 0.039 | 6.316 ± 0.393 |
3.368 ± 0.406 | 3.579 ± 0.694 |
5.474 ± 0.21 | 6.316 ± 3.339 |
8.842 ± 0.367 | 7.368 ± 1.44 |
2.105 ± 0.196 | 5.684 ± 0.419 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
