
Mycolicibacterium sp. P9-64
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycolicibacterium; unclassified Mycolicibacterium
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
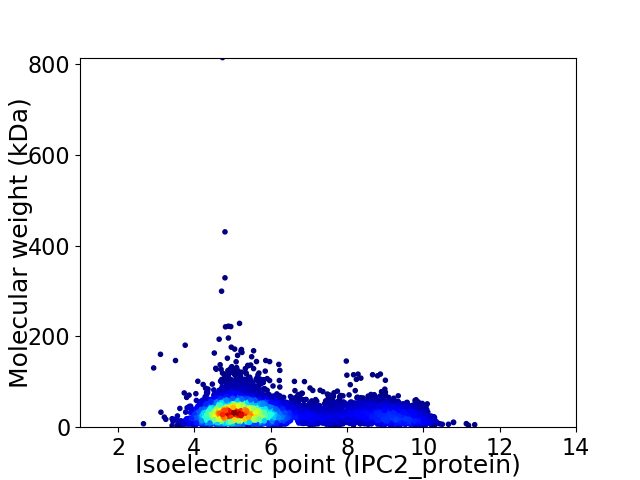
Virtual 2D-PAGE plot for 6799 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
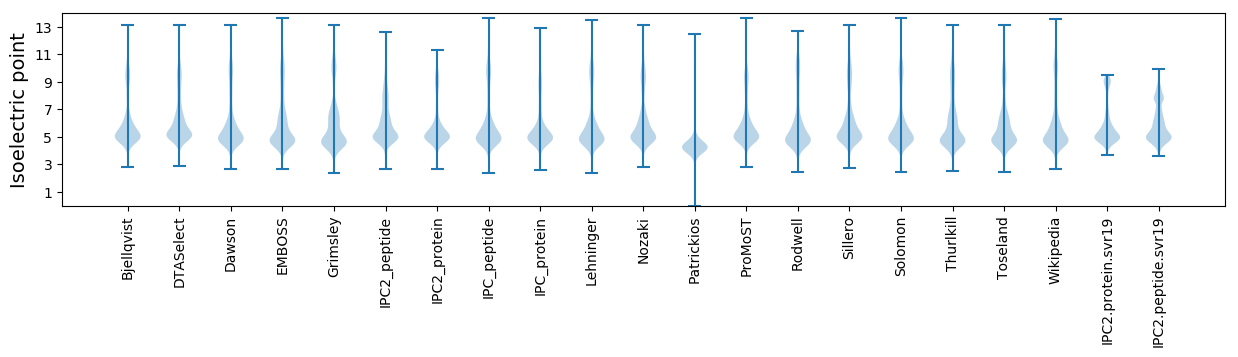
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5A7WRA0|A0A5A7WRA0_9MYCO MCE family protein OS=Mycolicibacterium sp. P9-64 OX=2024612 GN=CIW52_24080 PE=4 SV=1
MM1 pKa = 7.45VDD3 pKa = 3.61HH4 pKa = 6.25VQSGSADD11 pKa = 3.45KK12 pKa = 11.12VGSEE16 pKa = 4.22ASSTVSDD23 pKa = 3.59DD24 pKa = 3.37TVLRR28 pKa = 11.84RR29 pKa = 11.84SAITSAGQTASTTSGAQFSALSVPQVQTPPAPFDD63 pKa = 3.55PVGALLALPGQLFDD77 pKa = 3.6VASGFVALVITPFLAPTPNAPAQMPLLWTVLAWVRR112 pKa = 11.84RR113 pKa = 11.84EE114 pKa = 3.51ISHH117 pKa = 6.12TFFNRR122 pKa = 11.84SPIANPVQISQTVGGEE138 pKa = 3.96VTGNLNASDD147 pKa = 4.32PNGDD151 pKa = 3.58EE152 pKa = 4.07PLTYY156 pKa = 10.17TVTQQPAHH164 pKa = 5.54GTVVVRR170 pKa = 11.84PDD172 pKa = 2.95GTYY175 pKa = 10.06TYY177 pKa = 9.48TPTAGAPTSTLTDD190 pKa = 3.48SFTVTIDD197 pKa = 3.75DD198 pKa = 4.14SVGTQLPGVFGVVQGILHH216 pKa = 7.16RR217 pKa = 11.84LAQFIGIAQSDD228 pKa = 3.99TTVTQVAVTVAGAGVNLPPLVVTSVINPIPYY259 pKa = 8.89TVGSGPRR266 pKa = 11.84VLDD269 pKa = 3.22SHH271 pKa = 6.21LTVVDD276 pKa = 4.76GSSNLSGGTVSVGLGFSAGDD296 pKa = 3.66TLSYY300 pKa = 10.1TPPANNPITGSYY312 pKa = 10.39DD313 pKa = 2.99AATGVLTLSGTATVAQYY330 pKa = 11.14QDD332 pKa = 3.11ALRR335 pKa = 11.84AVSFSTTSDD344 pKa = 3.09ALIGARR350 pKa = 11.84TVSFVVTDD358 pKa = 3.9DD359 pKa = 4.46GGLSSISVPLALTVLALNLPPVVTTSVVGPIVTAGNPPVTLDD401 pKa = 3.63ANVTIIDD408 pKa = 4.11GSGSMTGAQVSLGLGFSAGDD428 pKa = 3.55TLGYY432 pKa = 7.62TAPQSNPITGSYY444 pKa = 9.68NAATGVLTLSGTGTVAQYY462 pKa = 10.84QEE464 pKa = 4.14ALRR467 pKa = 11.84AVTFSTTSNALVGVRR482 pKa = 11.84TVGFVVTDD490 pKa = 3.79DD491 pKa = 4.67GGLSSISVPLAVTVLALNLPPVVTTSVIGPIVTAGNPPVTLDD533 pKa = 3.57GNVTIVDD540 pKa = 4.48GSASMTGGTVSLGLGFSPGDD560 pKa = 3.52TLGYY564 pKa = 7.54TAPQNNPITGSYY576 pKa = 9.61NAATGVLTLSGTGTVAQYY594 pKa = 10.84QEE596 pKa = 4.14ALRR599 pKa = 11.84AVTFSTTSNALVGVRR614 pKa = 11.84TVGFVVTDD622 pKa = 3.79DD623 pKa = 4.67GGLSSISVPLAVTVLALNLPPVVTTSVIGPIVTAGNPPVTLDD665 pKa = 3.53ANVTIVDD672 pKa = 4.42GSASLTGGTVSLGLGFSAGDD692 pKa = 3.55TLGYY696 pKa = 7.61TAPQDD701 pKa = 3.69NPITGSYY708 pKa = 9.64NAATGVLTLSGTGTVAQYY726 pKa = 10.84QEE728 pKa = 4.14ALRR731 pKa = 11.84AVTFSTTSNALVGVRR746 pKa = 11.84TVGFVVTDD754 pKa = 3.79DD755 pKa = 4.67GGLSSISVPLAVTVLALNLPPVVTTSVVGPIVTAGNPPVTLDD797 pKa = 3.53ANVTIVDD804 pKa = 4.59GSASMTGGTVSLGLGFGPGDD824 pKa = 3.56TLGYY828 pKa = 7.54TAPQNNPITGSYY840 pKa = 9.61NAATGVLTLSGTGTVAQYY858 pKa = 10.84QEE860 pKa = 4.14ALRR863 pKa = 11.84AVTFSTTAGALVGVRR878 pKa = 11.84TVGFVVTDD886 pKa = 3.19VGGLSSISVPLAVTVLALNLPPVVTTSVIGPIVTAGNPPVTLDD929 pKa = 3.53ANVTIVDD936 pKa = 4.42GSASLTGGTVSVGLGFSAGDD956 pKa = 3.55TLGYY960 pKa = 7.6TAPQNNPITGSYY972 pKa = 10.39DD973 pKa = 2.99AATGVLTLSGTGTVAQYY990 pKa = 10.88QEE992 pKa = 4.36ALRR995 pKa = 11.84SITFSTTAGVLVGARR1010 pKa = 11.84TVSFVVTDD1018 pKa = 3.4VEE1020 pKa = 4.79GASSISVPLAVTVLALNLPPVVTTSVVGPIVTAGNPPVTLDD1061 pKa = 3.53ANVTIVDD1068 pKa = 4.42GSASLTGGTVSLGLGFSAGDD1088 pKa = 3.55TLGYY1092 pKa = 7.61TAPQDD1097 pKa = 3.69NPITGSYY1104 pKa = 10.4DD1105 pKa = 2.99AATGVLTLSGTGTVAQYY1122 pKa = 10.88QEE1124 pKa = 4.36ALRR1127 pKa = 11.84SITFSTTAGALVGARR1142 pKa = 11.84TVSFVVTDD1150 pKa = 3.4VEE1152 pKa = 4.79GASSISVPLAVTVLALNLPPVVTTSVVGPLVTAGNPPVTLDD1193 pKa = 3.53ANVTIVDD1200 pKa = 4.42GSASLTGGTVSLGLGFSAGDD1220 pKa = 3.55TLGYY1224 pKa = 7.61TAPQDD1229 pKa = 3.69NPITGSYY1236 pKa = 10.4DD1237 pKa = 2.99AATGVLTLSGTGTVAQYY1254 pKa = 10.88QEE1256 pKa = 4.36ALRR1259 pKa = 11.84SITFSTTAGALVGARR1274 pKa = 11.84TVSFVVTDD1282 pKa = 3.4VEE1284 pKa = 4.79GASSISVPLAVTVLALNLPPVVTTSVIGPIVTAGSPPVTLDD1325 pKa = 3.33ANVTIVDD1332 pKa = 4.42GSASLTGGTVSLGLGFSAGDD1352 pKa = 3.55TLGYY1356 pKa = 7.6TAPQNNPITGSYY1368 pKa = 10.71NSATGVLTLSGTGTVAQYY1386 pKa = 10.88QEE1388 pKa = 4.36ALRR1391 pKa = 11.84SITFSTTAGALVGARR1406 pKa = 11.84TVSFVVTDD1414 pKa = 3.4VEE1416 pKa = 4.79GASSISVPLAVTVLALNLPPVVTTSVIGPIVTAGNSPVTLDD1457 pKa = 3.57ANVTIVDD1464 pKa = 4.42GSASLTGGTVSVGLGFSAGDD1484 pKa = 3.55TLGYY1488 pKa = 7.61TAPQDD1493 pKa = 3.69NPITGSYY1500 pKa = 10.4DD1501 pKa = 2.99AATGVLTLSGTGTVAQYY1518 pKa = 10.88QEE1520 pKa = 4.36ALRR1523 pKa = 11.84SITFSTTAGALVGARR1538 pKa = 11.84TVSFVVTDD1546 pKa = 3.4VEE1548 pKa = 4.79GASSISVPLAVTVLALNVPPVVVTSILNLIPFTAGNAPATLDD1590 pKa = 3.69PNLTVIDD1597 pKa = 4.86GSASLSGATVAITVGLTAGDD1617 pKa = 3.93SLAFTQPPGSSITGSYY1633 pKa = 10.85NSATGVLTLAGTGTVAQYY1651 pKa = 10.85QEE1653 pKa = 4.29ALRR1656 pKa = 11.84SVTFATTTATLVGVRR1671 pKa = 11.84TVSFVVTDD1679 pKa = 3.58VEE1681 pKa = 4.5GLPSVSVPLAVTVALNLPPVVTSSVVGLSLNLLNDD1716 pKa = 4.63APPKK1720 pKa = 10.43ILDD1723 pKa = 3.96PGVLIVDD1730 pKa = 4.61DD1731 pKa = 4.91SSTLTGATVTVGGLVIGGNDD1751 pKa = 3.18TLAFTSPPGSGITGVWNAGTKK1772 pKa = 9.2TLTLSGAATVADD1784 pKa = 4.14YY1785 pKa = 8.47QTALRR1790 pKa = 11.84SVTFQTSGFLNLGARR1805 pKa = 11.84VVSFVVKK1812 pKa = 10.19DD1813 pKa = 3.54QQGQSSISVPLTVVVVGILL1832 pKa = 3.21
MM1 pKa = 7.45VDD3 pKa = 3.61HH4 pKa = 6.25VQSGSADD11 pKa = 3.45KK12 pKa = 11.12VGSEE16 pKa = 4.22ASSTVSDD23 pKa = 3.59DD24 pKa = 3.37TVLRR28 pKa = 11.84RR29 pKa = 11.84SAITSAGQTASTTSGAQFSALSVPQVQTPPAPFDD63 pKa = 3.55PVGALLALPGQLFDD77 pKa = 3.6VASGFVALVITPFLAPTPNAPAQMPLLWTVLAWVRR112 pKa = 11.84RR113 pKa = 11.84EE114 pKa = 3.51ISHH117 pKa = 6.12TFFNRR122 pKa = 11.84SPIANPVQISQTVGGEE138 pKa = 3.96VTGNLNASDD147 pKa = 4.32PNGDD151 pKa = 3.58EE152 pKa = 4.07PLTYY156 pKa = 10.17TVTQQPAHH164 pKa = 5.54GTVVVRR170 pKa = 11.84PDD172 pKa = 2.95GTYY175 pKa = 10.06TYY177 pKa = 9.48TPTAGAPTSTLTDD190 pKa = 3.48SFTVTIDD197 pKa = 3.75DD198 pKa = 4.14SVGTQLPGVFGVVQGILHH216 pKa = 7.16RR217 pKa = 11.84LAQFIGIAQSDD228 pKa = 3.99TTVTQVAVTVAGAGVNLPPLVVTSVINPIPYY259 pKa = 8.89TVGSGPRR266 pKa = 11.84VLDD269 pKa = 3.22SHH271 pKa = 6.21LTVVDD276 pKa = 4.76GSSNLSGGTVSVGLGFSAGDD296 pKa = 3.66TLSYY300 pKa = 10.1TPPANNPITGSYY312 pKa = 10.39DD313 pKa = 2.99AATGVLTLSGTATVAQYY330 pKa = 11.14QDD332 pKa = 3.11ALRR335 pKa = 11.84AVSFSTTSDD344 pKa = 3.09ALIGARR350 pKa = 11.84TVSFVVTDD358 pKa = 3.9DD359 pKa = 4.46GGLSSISVPLALTVLALNLPPVVTTSVVGPIVTAGNPPVTLDD401 pKa = 3.63ANVTIIDD408 pKa = 4.11GSGSMTGAQVSLGLGFSAGDD428 pKa = 3.55TLGYY432 pKa = 7.62TAPQSNPITGSYY444 pKa = 9.68NAATGVLTLSGTGTVAQYY462 pKa = 10.84QEE464 pKa = 4.14ALRR467 pKa = 11.84AVTFSTTSNALVGVRR482 pKa = 11.84TVGFVVTDD490 pKa = 3.79DD491 pKa = 4.67GGLSSISVPLAVTVLALNLPPVVTTSVIGPIVTAGNPPVTLDD533 pKa = 3.57GNVTIVDD540 pKa = 4.48GSASMTGGTVSLGLGFSPGDD560 pKa = 3.52TLGYY564 pKa = 7.54TAPQNNPITGSYY576 pKa = 9.61NAATGVLTLSGTGTVAQYY594 pKa = 10.84QEE596 pKa = 4.14ALRR599 pKa = 11.84AVTFSTTSNALVGVRR614 pKa = 11.84TVGFVVTDD622 pKa = 3.79DD623 pKa = 4.67GGLSSISVPLAVTVLALNLPPVVTTSVIGPIVTAGNPPVTLDD665 pKa = 3.53ANVTIVDD672 pKa = 4.42GSASLTGGTVSLGLGFSAGDD692 pKa = 3.55TLGYY696 pKa = 7.61TAPQDD701 pKa = 3.69NPITGSYY708 pKa = 9.64NAATGVLTLSGTGTVAQYY726 pKa = 10.84QEE728 pKa = 4.14ALRR731 pKa = 11.84AVTFSTTSNALVGVRR746 pKa = 11.84TVGFVVTDD754 pKa = 3.79DD755 pKa = 4.67GGLSSISVPLAVTVLALNLPPVVTTSVVGPIVTAGNPPVTLDD797 pKa = 3.53ANVTIVDD804 pKa = 4.59GSASMTGGTVSLGLGFGPGDD824 pKa = 3.56TLGYY828 pKa = 7.54TAPQNNPITGSYY840 pKa = 9.61NAATGVLTLSGTGTVAQYY858 pKa = 10.84QEE860 pKa = 4.14ALRR863 pKa = 11.84AVTFSTTAGALVGVRR878 pKa = 11.84TVGFVVTDD886 pKa = 3.19VGGLSSISVPLAVTVLALNLPPVVTTSVIGPIVTAGNPPVTLDD929 pKa = 3.53ANVTIVDD936 pKa = 4.42GSASLTGGTVSVGLGFSAGDD956 pKa = 3.55TLGYY960 pKa = 7.6TAPQNNPITGSYY972 pKa = 10.39DD973 pKa = 2.99AATGVLTLSGTGTVAQYY990 pKa = 10.88QEE992 pKa = 4.36ALRR995 pKa = 11.84SITFSTTAGVLVGARR1010 pKa = 11.84TVSFVVTDD1018 pKa = 3.4VEE1020 pKa = 4.79GASSISVPLAVTVLALNLPPVVTTSVVGPIVTAGNPPVTLDD1061 pKa = 3.53ANVTIVDD1068 pKa = 4.42GSASLTGGTVSLGLGFSAGDD1088 pKa = 3.55TLGYY1092 pKa = 7.61TAPQDD1097 pKa = 3.69NPITGSYY1104 pKa = 10.4DD1105 pKa = 2.99AATGVLTLSGTGTVAQYY1122 pKa = 10.88QEE1124 pKa = 4.36ALRR1127 pKa = 11.84SITFSTTAGALVGARR1142 pKa = 11.84TVSFVVTDD1150 pKa = 3.4VEE1152 pKa = 4.79GASSISVPLAVTVLALNLPPVVTTSVVGPLVTAGNPPVTLDD1193 pKa = 3.53ANVTIVDD1200 pKa = 4.42GSASLTGGTVSLGLGFSAGDD1220 pKa = 3.55TLGYY1224 pKa = 7.61TAPQDD1229 pKa = 3.69NPITGSYY1236 pKa = 10.4DD1237 pKa = 2.99AATGVLTLSGTGTVAQYY1254 pKa = 10.88QEE1256 pKa = 4.36ALRR1259 pKa = 11.84SITFSTTAGALVGARR1274 pKa = 11.84TVSFVVTDD1282 pKa = 3.4VEE1284 pKa = 4.79GASSISVPLAVTVLALNLPPVVTTSVIGPIVTAGSPPVTLDD1325 pKa = 3.33ANVTIVDD1332 pKa = 4.42GSASLTGGTVSLGLGFSAGDD1352 pKa = 3.55TLGYY1356 pKa = 7.6TAPQNNPITGSYY1368 pKa = 10.71NSATGVLTLSGTGTVAQYY1386 pKa = 10.88QEE1388 pKa = 4.36ALRR1391 pKa = 11.84SITFSTTAGALVGARR1406 pKa = 11.84TVSFVVTDD1414 pKa = 3.4VEE1416 pKa = 4.79GASSISVPLAVTVLALNLPPVVTTSVIGPIVTAGNSPVTLDD1457 pKa = 3.57ANVTIVDD1464 pKa = 4.42GSASLTGGTVSVGLGFSAGDD1484 pKa = 3.55TLGYY1488 pKa = 7.61TAPQDD1493 pKa = 3.69NPITGSYY1500 pKa = 10.4DD1501 pKa = 2.99AATGVLTLSGTGTVAQYY1518 pKa = 10.88QEE1520 pKa = 4.36ALRR1523 pKa = 11.84SITFSTTAGALVGARR1538 pKa = 11.84TVSFVVTDD1546 pKa = 3.4VEE1548 pKa = 4.79GASSISVPLAVTVLALNVPPVVVTSILNLIPFTAGNAPATLDD1590 pKa = 3.69PNLTVIDD1597 pKa = 4.86GSASLSGATVAITVGLTAGDD1617 pKa = 3.93SLAFTQPPGSSITGSYY1633 pKa = 10.85NSATGVLTLAGTGTVAQYY1651 pKa = 10.85QEE1653 pKa = 4.29ALRR1656 pKa = 11.84SVTFATTTATLVGVRR1671 pKa = 11.84TVSFVVTDD1679 pKa = 3.58VEE1681 pKa = 4.5GLPSVSVPLAVTVALNLPPVVTSSVVGLSLNLLNDD1716 pKa = 4.63APPKK1720 pKa = 10.43ILDD1723 pKa = 3.96PGVLIVDD1730 pKa = 4.61DD1731 pKa = 4.91SSTLTGATVTVGGLVIGGNDD1751 pKa = 3.18TLAFTSPPGSGITGVWNAGTKK1772 pKa = 9.2TLTLSGAATVADD1784 pKa = 4.14YY1785 pKa = 8.47QTALRR1790 pKa = 11.84SVTFQTSGFLNLGARR1805 pKa = 11.84VVSFVVKK1812 pKa = 10.19DD1813 pKa = 3.54QQGQSSISVPLTVVVVGILL1832 pKa = 3.21
Molecular weight: 180.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5A7WKF1|A0A5A7WKF1_9MYCO TetR/AcrR family transcriptional regulator OS=Mycolicibacterium sp. P9-64 OX=2024612 GN=CIW52_20695 PE=4 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVSGRR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.81GRR42 pKa = 11.84RR43 pKa = 11.84SLTAA47 pKa = 3.9
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVSGRR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.81GRR42 pKa = 11.84RR43 pKa = 11.84SLTAA47 pKa = 3.9
Molecular weight: 5.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2191175 |
27 |
7611 |
322.3 |
34.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.891 ± 0.035 | 0.792 ± 0.008 |
6.388 ± 0.024 | 5.123 ± 0.026 |
3.1 ± 0.017 | 8.943 ± 0.033 |
2.206 ± 0.014 | 4.382 ± 0.019 |
2.255 ± 0.018 | 9.777 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.059 ± 0.012 | 2.295 ± 0.015 |
5.688 ± 0.026 | 2.913 ± 0.016 |
6.883 ± 0.028 | 5.596 ± 0.019 |
6.216 ± 0.029 | 8.876 ± 0.028 |
1.477 ± 0.013 | 2.141 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
