
Papio ursinus cytomegalovirus
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Betaherpesvirinae; Cytomegalovirus; Cercopithecine betaherpesvirus 5
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
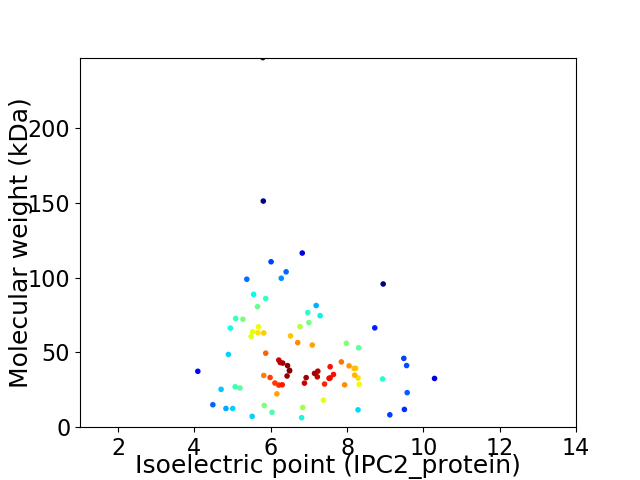
Virtual 2D-PAGE plot for 84 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
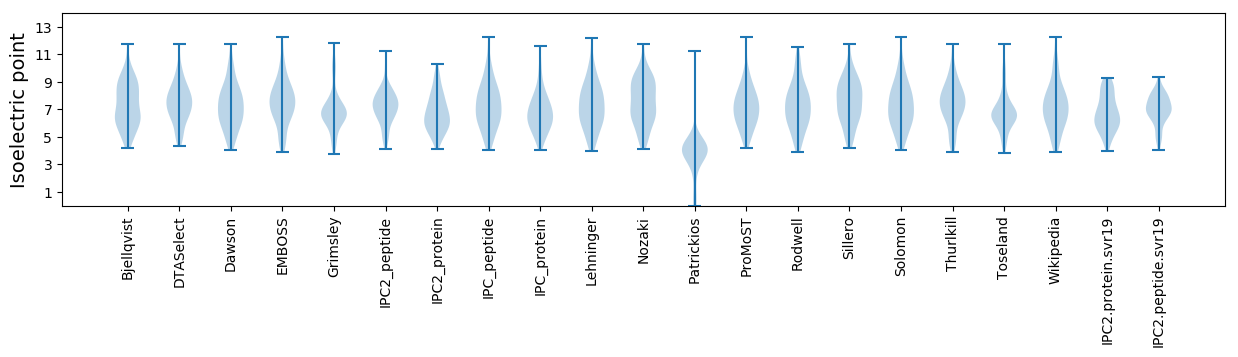
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F7G9A4|A0A0F7G9A4_SCMVC Envelope glycoprotein M OS=Papio ursinus cytomegalovirus OX=1667587 GN=gM PE=3 SV=1
MM1 pKa = 6.98QEE3 pKa = 4.3AMKK6 pKa = 9.44TWVLLVALFAITRR19 pKa = 11.84DD20 pKa = 3.74SACNNTSTASSSSTSSNTVTSSKK43 pKa = 11.27ANNTTSISSSTVSSTSPSGSSSATSNTTKK72 pKa = 10.2ATTSPATTNLSSASPASSPATATTNSTTTTPTPTSSKK109 pKa = 8.64NTSAGSSTPTNTSNKK124 pKa = 8.36TSPPASNTSTPSATKK139 pKa = 8.93TANSTTTNNSAAEE152 pKa = 4.13NTTNITPTFSTEE164 pKa = 3.67NVTVDD169 pKa = 3.38SNFTTGYY176 pKa = 10.24PNDD179 pKa = 4.08TFNATTDD186 pKa = 3.69PYY188 pKa = 11.3EE189 pKa = 3.99VMQIVDD195 pKa = 4.58LCNATISIIFTDD207 pKa = 3.44SDD209 pKa = 3.94EE210 pKa = 4.28EE211 pKa = 4.5TEE213 pKa = 4.32EE214 pKa = 4.37SDD216 pKa = 3.56EE217 pKa = 4.25QSSEE221 pKa = 4.01EE222 pKa = 3.98QQSFDD227 pKa = 4.84SIDD230 pKa = 3.71DD231 pKa = 3.58NTYY234 pKa = 9.62YY235 pKa = 10.16PVNPSYY241 pKa = 11.57SLLSDD246 pKa = 3.98SDD248 pKa = 3.77DD249 pKa = 3.57VMYY252 pKa = 9.61FYY254 pKa = 11.51ANCEE258 pKa = 4.18RR259 pKa = 11.84NDD261 pKa = 3.82TLSNTSCNYY270 pKa = 8.25TNQRR274 pKa = 11.84LTSWSLVSSVSFYY287 pKa = 10.52PAEE290 pKa = 3.91LTNCNKK296 pKa = 9.31PVAVIEE302 pKa = 4.33VGNSSLVVSAEE313 pKa = 3.94ATSNLVDD320 pKa = 4.94GIYY323 pKa = 10.47KK324 pKa = 10.24VLGVPDD330 pKa = 4.63LNSSFLKK337 pKa = 10.55EE338 pKa = 3.63LAKK341 pKa = 10.32FQQLIIQGQNKK352 pKa = 7.72PQKK355 pKa = 10.32
MM1 pKa = 6.98QEE3 pKa = 4.3AMKK6 pKa = 9.44TWVLLVALFAITRR19 pKa = 11.84DD20 pKa = 3.74SACNNTSTASSSSTSSNTVTSSKK43 pKa = 11.27ANNTTSISSSTVSSTSPSGSSSATSNTTKK72 pKa = 10.2ATTSPATTNLSSASPASSPATATTNSTTTTPTPTSSKK109 pKa = 8.64NTSAGSSTPTNTSNKK124 pKa = 8.36TSPPASNTSTPSATKK139 pKa = 8.93TANSTTTNNSAAEE152 pKa = 4.13NTTNITPTFSTEE164 pKa = 3.67NVTVDD169 pKa = 3.38SNFTTGYY176 pKa = 10.24PNDD179 pKa = 4.08TFNATTDD186 pKa = 3.69PYY188 pKa = 11.3EE189 pKa = 3.99VMQIVDD195 pKa = 4.58LCNATISIIFTDD207 pKa = 3.44SDD209 pKa = 3.94EE210 pKa = 4.28EE211 pKa = 4.5TEE213 pKa = 4.32EE214 pKa = 4.37SDD216 pKa = 3.56EE217 pKa = 4.25QSSEE221 pKa = 4.01EE222 pKa = 3.98QQSFDD227 pKa = 4.84SIDD230 pKa = 3.71DD231 pKa = 3.58NTYY234 pKa = 9.62YY235 pKa = 10.16PVNPSYY241 pKa = 11.57SLLSDD246 pKa = 3.98SDD248 pKa = 3.77DD249 pKa = 3.57VMYY252 pKa = 9.61FYY254 pKa = 11.51ANCEE258 pKa = 4.18RR259 pKa = 11.84NDD261 pKa = 3.82TLSNTSCNYY270 pKa = 8.25TNQRR274 pKa = 11.84LTSWSLVSSVSFYY287 pKa = 10.52PAEE290 pKa = 3.91LTNCNKK296 pKa = 9.31PVAVIEE302 pKa = 4.33VGNSSLVVSAEE313 pKa = 3.94ATSNLVDD320 pKa = 4.94GIYY323 pKa = 10.47KK324 pKa = 10.24VLGVPDD330 pKa = 4.63LNSSFLKK337 pKa = 10.55EE338 pKa = 3.63LAKK341 pKa = 10.32FQQLIIQGQNKK352 pKa = 7.72PQKK355 pKa = 10.32
Molecular weight: 37.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F7G9M3|A0A0F7G9M3_SCMVC UL74A OS=Papio ursinus cytomegalovirus OX=1667587 PE=4 SV=1
MM1 pKa = 6.56WHH3 pKa = 6.75AAILLYY9 pKa = 10.65EE10 pKa = 4.2PTLTASAALALGNAISAIHH29 pKa = 6.31HH30 pKa = 6.11RR31 pKa = 11.84FSMLCARR38 pKa = 11.84CRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84PATEE46 pKa = 3.64SDD48 pKa = 3.81SNNARR53 pKa = 11.84SSQSEE58 pKa = 3.77ASQRR62 pKa = 11.84RR63 pKa = 11.84QTRR66 pKa = 11.84EE67 pKa = 3.68NQKK70 pKa = 10.4EE71 pKa = 3.87PSPKK75 pKa = 10.26NNNTSLVGRR84 pKa = 11.84ALVIIGQQLIAANLGACPPPVTLFRR109 pKa = 5.98
MM1 pKa = 6.56WHH3 pKa = 6.75AAILLYY9 pKa = 10.65EE10 pKa = 4.2PTLTASAALALGNAISAIHH29 pKa = 6.31HH30 pKa = 6.11RR31 pKa = 11.84FSMLCARR38 pKa = 11.84CRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84PATEE46 pKa = 3.64SDD48 pKa = 3.81SNNARR53 pKa = 11.84SSQSEE58 pKa = 3.77ASQRR62 pKa = 11.84RR63 pKa = 11.84QTRR66 pKa = 11.84EE67 pKa = 3.68NQKK70 pKa = 10.4EE71 pKa = 3.87PSPKK75 pKa = 10.26NNNTSLVGRR84 pKa = 11.84ALVIIGQQLIAANLGACPPPVTLFRR109 pKa = 5.98
Molecular weight: 11.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
36678 |
56 |
2183 |
436.6 |
49.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.271 ± 0.247 | 2.576 ± 0.153 |
5.128 ± 0.189 | 5.314 ± 0.18 |
4.313 ± 0.129 | 4.681 ± 0.206 |
3.065 ± 0.126 | 4.774 ± 0.225 |
3.757 ± 0.227 | 10.333 ± 0.24 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.487 ± 0.105 | 3.915 ± 0.185 |
5.466 ± 0.224 | 3.888 ± 0.2 |
6.68 ± 0.261 | 7.539 ± 0.265 |
6.584 ± 0.287 | 7.271 ± 0.189 |
1.279 ± 0.093 | 3.678 ± 0.118 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
